Abstract
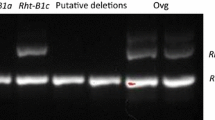
Overlapping nucleotides or overgos were designed and used to detect deletions in rice mutants that had been generated by treatment with a chemical (diepoxybutane, DB) or irradiation (gamma ray, GR). As a proof of concept, DNA of three spotted leaf 11 (spl11) deletion mutants, GR5612, GR5717 and DB2487, and the wild-type DNA (IR64) were spotted onto nylon membranes in different concentrations and probed with radiolabeled overgos designed from different regions within the Spl11 gene. At least 3 μg per spot of DNA was required to show unambiguous and visible signals. Overgos designed from regions putatively deleted in GR5612 consistently did not hybridize to the GR5612 genomic DNA, whereas DNA from wild-type IR64 and from mutants in other regions consistently hybridized to the overgos. DNA of mutant DB2487 hybridized to all the overgos tested, suggesting the deletion is small and undetectable by the overgos. Overall, this study demonstrates that overgos can be employed to detect, verify, and characterize deletions, particularly in single copy genes, in mutant genomes. However, the technique requires careful adjustment of DNA concentration before spotting and the spotting of relatively high DNA concentrations onto the membranes.




Similar content being viewed by others
Abbreviations
- Overgos:
-
overlapping nucleotides
- Spl11 :
-
spotted leaf 11
- DB:
-
diepoxybutane
- GR:
-
gamma ray
References
Aguirrezabalaga I, Nivard M, Comendador M, Vogel E. The cross-linking agent hexamethylyphosphoramide predominantly induces intra-locus and multi-locus deletions in postmeiotic germ cells of Drosophila. Genetics 1995;139:649–58.
Anderson P, Okubara P, Arroyo-Garcia R, Meyers B, Michelmore R. Molecular analysis of irradiation-induced and spontaneous deletion mutants at a disease resistance locus in Lactuca sativa. Mol Gen Genet 1996;251:316–25.
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (editors) Current protocols in molecular biology, supplement 21, pp 2.9.1–2.9.6. Wiley, New York; 1993
Bouchez D, Hofte H. Functional genomics in plants. Plant Physiol 1998;118:725–32.
Doyle JJ, Doyle JL. A rapid total DNA preparation procedure for fresh plant tissue. Focus 1990;12:13–5.
Edgley M, D’Souza A, Moulder G, McKay S, Shen B, Gilchrist E, et al. Improved detection of small deletions in complex pools of DNA. Nucleic Acid Res 2002;30:12e52.
Feng J, Vick BA, Lee MK, Zhang HB, Jan CC. Construction of BAC and BIBAC libraries from sunflower and identification of linkage group-specific clones by overgo hybridization. Theor Appl Genet 2006;113:23–32.
Gardiner J, Schroeder S, Polacco ML, Villeda HS, Fang Z, Morgante M, et al. Anchoring 9,371 maize expressed sequence tagged unigenes to the bacterial artificial chromosome contig map by two-dimensional overgo hybridization. Plant Physiol 2004;134:1317–26.
Han CS, Sutherland RD, Jewett PB, Campbell ML, Meincke LJ, Tesmer JG, et al. Construction of a BAC contig map of chromosome 16q by two-dimensional overgo hybridization. Genome Res 2000;10:714–21.
Li X, Song Y, Century K, Straight S, Ronald P, Dong X, et al. A fast neutron deletion mutagenesis-based reverse genetics system for plants. Plant J 2001;27:235–42.
Liu LX, Spoerke JM, Mulligan EL, Chen J, Reardon B, Westlund B, et al. High-throughput isolation of Caenorhabditis elegans deletion mutants. Genome Res 2000;9:859–67.
Manosalva P, Ryba-White M, Wu C, Lei C, Baraoidan M, Leung H, et al. A PCR-based screening strategy for detecting deletions in defense response genes in rice. Phytopathology 2003;93:S57.
Martienssen RA. Functional genomics: probing plant gene function and expression with transposons. Proc Natl Acad Sci U S A 1998;95:2021–26.
Reardon J, Liljestrand-Golden C, Dusenbery R, Smith P. Molecular analysis of diepoxybutane-induced mutations at the rosy locus of Drosophila melanogaster. Genetics 1987;115:323–31.
Ross MT, LaBrie S, McPherson J, Stanton VP. Screening of large-insert libraries by hybridization. In: Dracopoli NC, Haines JL, Korf DT, Moir CC, Morton CC, Seidman CE, Seidman JG, Smith DR (editors), Current protocols in human genetics, pp. 5.6.1–5.6.52. Wiley, New York; 1999
Shirley BS, Hanley S, Goodman HM. Effects of ionizing radiation on a plant genome: analysis of two Arabidopsis transparent testa mutations. Plant Cell 1992;4:333–47.
Song BK, Nadarajah K, Romanov MN, Ratnam W. Cross-species bacterial artificial chromosome (BAC) library screening via overgo-based hybridization and BAC-contig mapping of a yield enhancement quantitative trait loci (QTL) YLD1.1 in the Malaysian wild rice Oryza rufipogon. Cell Mol Biol Lett 2005;10:425–37.
Thorsen J, Zhu B, Frengen E, Osoegawa K, de Jong PJ, Koop BF, et al. A highly redundant BAC library of Atlantic salmon (Salmo salar): an important tool for salmon projects. BMC Genomics 2005;6:50–7.
Wu JL, Wu C, Lei C, Baraoidan M, Bordeos A, Madamba MRS, et al. Chemical- and irradiation-induced mutants of indica rice IR64 for forward and reverse genetics. Plant Mol Biol 2005;59:85–97.
Yandell MD, Edgar LG, Wood WB. Trimethylpsoralen induces small deletion mutations in Caenorhabditis elegans. Proc Natl Acad Sci U S A 1994;91:1381–85.
Yuksel B, Paterson AH. Construction and characterization of a peanut HindIII BAC library. Theor Appl Genet 2005;111:630–9.
Yuksel B, Estill JC, Schulze SR, Paterson AH. Organization and evolution of resistance gene analogs in peanut. Mol Gen Genomics 2005a;274:248–63.
Yuksel B, Bowers JE, Estill J, Goff L, Lemke C, Paterson AH. Exploratory integration of peanut genetic and physical maps and possible contributions from Arabidopsis. Theor Appl Genet 2005b;111:87–94.
Yin Z, Chen J, Zeng L, Goh M, Leung H, Khush GS, et al. Characterizing rice lesion mimic mutants and identifying a mutant with broad-spectrum resistance to rice blast and bacterial blight. Mol Plant Microbe Interact 2000;13:869–76.
Zeng LR, Qu S, Bordeos A, Yang C, Baraoidan M, Yan H, et al. Spotted leaf 11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 2004;16:2795–808.
Acknowledgments
MGQ Diaz is a recipient of a Postdoctoral Grant from the University of the Philippines System Faculty Development Program. The authors would like to thank Dr. Guo-Liang Wang for providing the spl11 mutant seeds and the sequences of the presumed deleted segments. This work is supported by the Generation Challenges Program, USDA-CSREES-NRI, and the USAID-IRRI linkage program.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Diaz, M.G.Q., Ryba, M., Leung, H. et al. Detection of Deletion Mutants in Rice Via Overgo Hybridization Onto Membrane Spotted Arrays. Plant Mol Biol Rep 25, 17–26 (2007). https://doi.org/10.1007/s11105-007-0002-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-007-0002-7