Abstract
Two swine paramyxoviruses (SPMV)—(81-19252 (Texas-81) and 92-7783 (ISU-92)—were isolated from encephalitic pigs in the United States in 1981 and 1992. Antigenic, morphologic, and biological characteristics of these two viruses were essentially similar to members of the family Paramyxoviridae. Antigenic analysis by indirect fluorescent antibody, immunoblot, and one-way cross-neutralization tests placed these viruses along with bovine parainfluenza 3 (BPIV3) viruses. Purified virions were 50–300 nm in size and morphologically indistinguishable from other paramyxoviruses. These two viruses hemagglutinated red blood cells and had neuraminidase activity. The gene junctions of fusion (F) and hemagglutinin (HN) glycoprotein genes of these viruses contained highly conserved transcription start and stop signal sequences and trinucleotide intergenic regions similar to other Paramyxoviridae. The F gene of ISU-92 was longer than Texas-81 due to insertion of a 24-nucleotide “U”-rich 3′ untranslated region. Structure-based sequence alignment of glycoproteins of these two SPMVs indicated that they are essentially similar in structure and function to parainfluenzaviruses. The Texas-81 strain was closely related to BPIV3 Shipping Fever (SF) strain at nucleotide and amino acid level, while the ISU-92 strain was more closely related to BPIV3 910N strain. The envelope glycoproteins of ISU-92 had only ~92 and ~96% identity at nucleotide and amino acid levels with BPIV3-SF strain, respectively. The high sequence identities to BPIV3 indicated cross-species infection in pigs. Phylogenetic analyses based on both F protein and HN protein suggested the classification of these viruses into the subfamily Paramyxovirinae, genus Respirovirus, and genotype A of BPIV3.
Similar content being viewed by others
Introduction
Paramyxoviruses are established pathogens of the central nervous and respiratory systems in many host species. In the last few decades, many novel paramyxoviruses have emerged causing catastrophic illnesses in different aquatic and terrestrial species of animals and some of them also made the species jump to humans. Members of the family Paramyxoviridae are enveloped viruses possessing a nonsegmented negative-strand genome and are divided into two subfamilies, Paramyxovirinae and Pneumovirinae. Currently, there are five genera within the subfamily Paramyxovirinae: Rubulavirus, Avulavirus, Respirovirus, Morbillivirus, and Henipavirus [1]. La Piedad Michoacan paramyxovirus (LPMV) is the only well-studied neurotropic paramyxovirus isolated from pigs prior to the 1990s. LPMV was first isolated in central Mexico in the early 1980s [2], and it has become endemic in Mexico [3]. This virus induced interstitial pneumonia and encephalitis in pigs. There were extensive records of paramyxoviruses derived from the brain or nasal swabs of sick pigs in Japan, in 1950s [4], Canada, in 1971 [5], in Israel, in 1986 [6] as well as the United States, in 1960s and 1980s in Texas [6]. There was also concurrent infection of porcine reproductive and respiratory syndrome virus and a paramyxovirus in Germany in the 1990s [7] that has been subsequently named “SER” virus [8].
Since 1994, four bat-associated paramyxoviruses have emerged, three of which caused disease in animals and humans [9]. In late 1994 in Australia, Hendra virus (HeV) caused an outbreak of severe respiratory disease resulting in the death of 13 horses and their trainer [10], followed by sporadic HeV outbreaks in horses and humans [11]. A closely related virus, Nipah virus (NiV) from Malaysia, caused severe febrile encephalitis and death in pigs and humans [12], which spread to Bangladesh and India [13, 14]. In 1997, another paramyxovirus, named Menangle virus (MenV), has been isolated in Australia from still-born pigs with deformities [4], and associated human illness [15]. In 2000, Tioman virus (TioPV) was isolated from urine collected beneath a fruit bat colony on Tioman Island, Malaysia [16]. Molecular characterization revealed that MenV and TiV are closely related novel members of the genus Rubulavirus [16–18]. Mapuera virus (MPRV) was isolated in 1979 from the salivary glands of an apparently healthy fruit bat (Sturnira lilium), captured in the tropical rainforest of Brazil [19]. J-virus (JV), isolated from wild mice in Australia, and Beilong virus (BeV), originally isolated from human mesangial cells in China and subsequently detected in rat mesangial cells, represent a new group of paramyxoviruses [20–24]. Recently, novel paramyxoviruses were also isolated from Atlantic bottlenose dolphins and Atlantic salmons and characterized [25, 26].
Two swine paramyxoviruses (SPMV)—(81-19252 (Texas-81) and 92-7783 (ISU-92))—were isolated from the brain of each pig that experienced respiratory and central nervous system disease in the 1980s and 1990s from South and North Central United States, respectively. ISU-92 virus was isolated from a swine operation in the 1990s in north-central United States. The outbreak started in a continuous-flow finishing barn of 400 pigs with signs of respiratory disease. Affected pigs showed high fever and mild cough, and 17 pigs died within 4 days. Ten days later, the epizootic spread to a second finishing barn and affected all pigs of different ages. Encephalitic signs were observed from several pigs with persistent squealing, head pressing, whole-body tremors, and hind-limb ataxia [6]. Texas-81 virus was isolated from the brain of each pig that exhibited respiratory and neurological disease in Texas State, in 1981. Further information on the history of this virus is not available. Antigenic analysis by indirect fluorescent antibody assay (IFA) at the National Veterinary Services Laboratory (NVSL), Ames, IA indicated that these two viruses are closely related to human parainfluenza virus (HPIV) type 1 and 3 and bovine parainfluenza virus type 3 (BPIV3).
As new viruses emerge that cause severe diseases in pigs and have the potential to cross species barrier and infect other hosts including humans, it is important that these new viruses need to be studied further. Although paramyxoviruses have been isolated from swine in United States and other parts of the world, their importance as swine pathogens or zoonotic agents remains to be determined. To understand the significance of these two strains of SPMV for swine health and to determine their taxonomic status, we have initiated molecular characterization studies. Here, we describe the biological and molecular characterization of the fusion (F) and hemagglutinin (HN) glycoprotein genes and their phylogenetic identity with other paramyxoviruses.
Materials and methods
Viruses and cells
Texas-81 and ISU-92 were received from the NVSL, Animal and Plant Health Inspection Service (APHIS), United States Department of Agriculture (USDA), Ames, IA. Porcine Kidney (PK15) cells, tested free of porcine circovirus (PCV), were obtained from Dr. X. J. Meng, Virginia Polytechnic Institute and State University, and used to propagate these swine paramyxoviruses. Vero cells (ATCC #C1008) were used for fusion and plaque assays.
Virus purification
Iodixanol gradients (OptiPrep, Sigma) were prepared in phosphate-buffered saline (PBS) (14–26%). Cell lysates from virus stock (plaque purified) were layered onto the top of the gradient and centrifuged for 1.5 h at 250,000×g in a SW41 Ti rotor. Virus fraction was collected from the gradient and was either examined by transmission electron microscopy or analyzed for protein content by immunoblot.
Transmission electron microscopy
Purified virus was adsorbed to glow discharged carbon films on 400-mesh grids. The grids were then washed in PBS and fixed with 2% paraformaldehyde (10 min) and extensively washed with distilled water before negatively stained with 1% phosphotungstic acid. All specimens were observed in Philips EM 420 transmission electron microscope operating at 100 kV.
Plaque assay
Confluent Vero cells in 6-well plates were infected with virus stocks prepared from the third limiting dilution passage in PK15 cells. Viruses were diluted in Dulbecco’s Modified Eagle Medium (DMEM) (Invitrogen), incubated at 37°C in a 5% CO2 incubator for 1 h, washed with PBS, and then overlaid with 0.8% methyl cellulose in DMEM with 2% FBS. The cells were fixed with methanol–acetone (1:1) at 6 days post-infection (dpi) and stained with 1% crystal violet. The mean number of plaque forming units (pfu) ml−1 for each virus was determined.
Hemagglutination (HA) assay
Swine red blood cells (sRBCs) were washed three times with PBS and resuspended in PBS as a 1% suspension. Texas-81 and ISU-92 virus stocks prepared in Vero cells were titrated in doubling dilutions against sRBC in V-bottom microtiter plates (Nunc).
Fusion index assay
The fusogenicity of Texas-81 and ISU-92 viruses was examined as described by Kohn and Fuchs [27]. Viruses were inoculated into confluent Vero cells in 6-well plates at a MOI of 0.1. Cells were maintained in DMEM with 2% FBS at 37°C in a 5% CO2 environment. After 72 h, the medium was removed, and cells were washed once with 0.02% EDTA and then incubated with 1 ml of 0.02% EDTA for 2 min at room temperature. The cells were then washed with PBS and fixed with methanol and stained with hematoxylin-eosin (Hema 3, Sigma). Fusion was quantitated by expressing the fusion index as the ratio of the total number of nuclei to the number of cells in which these nuclei were observed (i.e., the mean number of nuclei per cell). The formation of syncytia was visualized by staining virus-infected cells at 48 h with Hema 3 without EDTA treatment.
Fusogenicity of Texas-81 and ISU-92 F genes were also tested in a plasmid-based system. The open reading frames of the F and HN genes of Texas-81 and ISU-92 viruses were cloned into pCAGGS plasmid and transfected into Vero cells (1 μg of each plasmid) using Lipofectamine 2000 (Invitrogen). At 72 h post-transfection, fusion index was calculated as described above.
Neuraminidase (NA) activity
The NA activity of the Texas-81 and ISU-92 strains was determined by a fluorescence-based NA assay according to the procedures of Potier et al. [28, 29]. Newcastle Disease Virus Beaudette C strain was used as a standard control virus. Briefly, serial dilutions of virus strains in dilution buffer (32.5 mM MES (2-(N-morpholino) ethanesulfonic acid, sodium salt, Sigma–Aldrich) pH 5.8, 4 mM CaCl2) were prepared in a 96-well black flat bottom plates. Then, the same volume of substrate buffer (2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid, sodium salt (MUN, Sigma–Aldrich) prepared in 32.5 mM MES pH 5.8, 4 mM CaCl2 buffer) was added to each well. The plate was gently shaken on a mechanical vibrator and then incubated for 1 h at 37°C. The reaction was terminated by adding 150 μl of 50 mM glycine buffer pH 10.4. The results were read in a VIKTOR multilabel reader (TECAN Safire2) with an excitation wavelength of 360 nm and an emission wavelength of 450 nm.
IFA test
Virus stocks were inoculated into confluent Vero cells in 8-well chambers at a MOI of 1.0, 0.1, and 0.01. Cells were maintained in DMEM with 2% FBS at 37°C in a 5% CO2 environment for 48 h and observed for cytopathic effects. The cells were then washed with PBS and fixed with methanol–acetone followed by three washes with PBST (0.1% Tween 20 in PBS). The cell monolayers were blocked overnight at 4°C with PBST containing 5% skim milk powder (PBSM). After three brief washes with PBST, the cells were incubated with antibovine PIV3 polyclonal antibody (NVSL) diluted 1:100 in PBSM and incubated overnight at 4°C. Then, the cells were washed three times with PBST and incubated with FITC-antibovine IgG (KPL) (1:32 dilution in PBSM) at 37°C in a moist environment for 1 h. After three washes with PBST, the slides were layered with glycerol–PBS (1:1) and observed under a Nikon Eclipse TS-100 epifluorescence microscope.
Immunoblot
OptiPrep-purified virions were subjected to immunoblotting using standard procedures. Briefly, the proteins were separated in a 4–20% sodium-dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to Immobilon-P (Millipore) membranes. The viral proteins were probed with antibovine PIV3 polyclonal bovine antibody (NVSL) (1:100 dilution), followed by HRP-labeled antibovine IgG (KPL) (1:500 dilution) secondary antibodies. The blots were visualized with chemiluminescent ECL Western blotting system (GE Healthcare).
One-way cross-neutralization test
Monospecific convalescent sera were obtained from pigs inoculated intranasally with 5 × 107 TCID50 of Texas-81 or ISU-92 virus stock. One-way cross-neutralization test was performed in 96-well plates against 500 TCID50 of respective viruses and heat-inactivated twofold diluted sera. The cells were fixed on day 5 and stained with Hema 3 to determine the virus-neutralization titer.
Reverse transcription (RT)-polymerase chain reaction (PCR) and sequencing
RNA extraction and RT reactions were performed using standard procedures. Briefly, virus-infected PK15 cells were scraped into the medium and subjected to three cycles of freezing and thawing. After initial clarification at 3,000×g for 15 min, polyethylene glycol 8000 (Sigma) was added to the cell lysate to a concentration of 10% and the lysate was incubated for 4 h at 4°C. The virus was pelleted at 12,000×g for 60 min at 4°C, and the viral genomic RNA was extracted from the virus pellet using RNeasy Mini Kit (QIAgen). Oligonucleotide primers were designed based on consensus bovine parainfluenza virus 3 (BPIV3) F and HN gene nucleotide sequences (primer sequences available upon request). The cDNA copies of the genomic RNA of the two virus strains were synthesized using the specific oligonucleotide primers and Superscript III reverse transcriptase (Invitrogen). Subsequently, Platinum Taq DNA Polymerase (Invitrogen) was used to amplify the F or HN gene. The PCR products were purified using Qiaquick PCR purification kit (Qiagen). The PCR products were cloned using the TA cloning system (Invitrogen). Both purified PCR products and at least 10 TA clones were sequenced in both directions in an automated sequencer (Core Laboratory Facility at Virginia Bioinformatics Institute, Virginia Tech). Based on the sequencing results, primer-walking strategy was employed, and new sets of oligonucleotide primers were designed for sequencing the middle part of the genes.
Determination of antigenic relatedness
The antigenic relatedness of SPMV strains were expressed by R value, calculated using the following formula of Archetti and Horsfall [30].
where r 1 is the ratio of the heterologous titer obtained with virus 2 to the homologous titer obtained with virus 1; r 2 is the ratio of the heterologous titer obtained with virus 1 to the homologous titer obtained with virus 2; and R is a geometric mean of ratio r 1 and r 2, which is used to express the antigenic relatedness between two viruses when both antigens and antisera were used in indirect FA test.
R values were interpreted by the method of Brooksby [31] for similarity comparison of two viruses. The criteria are as follows: (1) R value of 1 or close to 1 indicates antigenic identity between two tested viruses; (2) R value >70% means little or no difference between two viruses tested; (3) R value between 33 and 70% indicates a minor subtype difference; (4) R value between 11 and 32% represents a major subtype difference; and (5) R value between 0 and 10% represents a serotype difference.
Data analysis
Nucleotide sequence editing, alignment, and prediction of amino acid sequence and analyses were conducted using the software package DNASTAR (Lasergene). Phylogenetic relationships of these two viruses within Paramyxoviridae family were constructed by Phylogenetic Analysis using Parsimony (PAUP 4.01) software with 1,000 bootstrap replicates. The protein functional analyses of deduced amino acid sequences were carried out using online resources available at ExPASy Proteomics tools (http://us.expasy.org/tools/).
Accession numbers for sequence analyses
The sequences of ISU-92 and Texas-81 were deposited in GenBank under accession # EU439428 and EU439429, respectively. The accession numbers for other viral sequences used for phylogenetic analysis are: Atlantic salmon paramyxovirus (ASPV), EF646380; Avian metapneumovirus (AMPV), NC_007652; Avian paramyxovirus 6 (APMV6), NC_003043; Beilong virus, NC_007803; J-virus, NC_007454; Bovine parainfluenza virus 3-910N (BPIV3-910N), D84095; Bovine parainfluenza virus 3 strain Kansas/15626/84 (BPIV3-Ka), AF178654; Bovine parainfluenza virus Q5592 (BPIV3-Q5592), EU277658; Bovine parainfluenza virus 3 strain Shipping Fever (BPIV3-SF), AF178655; Bovine parainfluenza virus 3 strain SK-217 (BPIV3-SK217), U31671; Bovine respiratory syncytial virus (BRSV), NC_001989; Canine distemper virus (CDV), NC_001921; Dolphin morbillivirus (DMV), NC_005283; Fer-de-lance virus (FDLV), NC_005084; Hendra virus (HeV), AF017149; Human metapneumovirus (HMPV), NC_004148; Human parainfluenza virus 1 strain Washington/1964 (HPIV1-Wa), NC_003461; Human parainfluenza virus 2 (HPIV2), NC_003443; Human parainfluenza virus 3 (HPIV3), AB012132; Human parainfluenza virus 3-GPv (HPIV3-GPv), NC_001796; Human parainfluenza virus 3-JS (HPIV3-JS), Z11575; Human respiratory syncytial virus (HRSV), NC_001781; J-virus, NC_007454; Measles virus (MV), NC_001498; Menangle virus (MeNV), NC_007620; Mossman virus (MoV), NC_005339; Mumps virus (MuV), NC_002200; Newcastle disease virus (NDV), NC_002617; Nipah virus (NiV), NC_002728; Peste-des-petits-ruminants virus (PPRV), NC_006383; Pneumonia virus of mice, AY573818; Porcine Rubulavirus (PoRV/LPMV), NC_009640; Rinderpest virus (RPV) (strain Kabete O), NC_006296; Sendai virus (SeV), NC_001552; Simian parainfluenza virus 5 (SV5), NC_006430; Tioman virus (TioPV), NC_004074; and Tupaia paramyxovirus (TMPV), NC_002199.
Results and discussion
Morphological and biological properties
Transmission electron microscopy revealed spherical to pleomorphic virions approximately 50–300 nm in diameter morphologically indistinguishable from paramyxoviruses [32]. Intact virions were enveloped and densely packed with surface projections representing viral glycoprotein spikes. Nucleocapsids were visible and exhibited a typical “herringbone” pattern (Fig. 1). Both Texax-81 and ISU-92 viruses were able to agglutinate sRBCs. The HA titers of Texas-81 and ISU-92 were 24 and 25, respectively. The agglutinated sRBCs eluted after 1 h suggesting neuraminidase function. Neuraminidase assay also confirmed this. BPIV3 has been shown to have hemagglutinating and neuraminidase functions [33]. The fusion index of Texas-81 was 63 nuclei per cell and of ISU-92 was 60 nuclei per cell (72 hpi, MOI, 0.1), indicating that these viruses are highly fusogenic (Fig. 2). The fusion index with Texas-81 and ISU-92 F plasmids with homologous HN plasmids was 22.7 and 13.5, respectively. The viruses were able to form plaques in Vero cells at 6 dpi and grew to 2.62 × 107 pfu ml−1 (Texas-81) and 5.75 × 107 pfu ml−1 (ISU-92). Both viruses produced similar-sized plaques in Vero cells (Fig. 2).
Ultrastructure of swine paramyxoviruses. Purified virions were negatively stained with 1% phosphotungstic acid and viewed under a transmission electron microscope. a, c ISU-92 virus. b, d Texas-81 virus. Intact virion with fine surface projections (arrow a) representing the viral glycoprotein spikes. Highly pleomorphic viral particles with extruded nucleocapsids were indicated by arrow b. Nucleocapsids exhibit a typical “herringbone” pattern
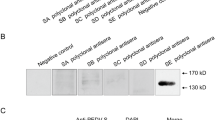
a–c Syncytium formation in Vero cells by Texas-81 and ISU-92. The fusogenicity of Texas-81 and ISU-92 viruses was examined in Vero cells. a ISU-92. b Texas-81. c Mock-infected Vero cells. d–f Plaque assay. Confluent Vero cells in 6-well plates were infected with serially diluted virus stocks and then overlaid with 0.8% methyl cellulose in DMEM with 2% FBS. Plaque sizes of Texas-81 and ISU-92 viruses were similar. d ISU-92. e Texas-81. f Mock infected. g–j Antigenic analysis of swine paramyxoviruses. g–i Indirect fluorescence antibody assay was performed by infecting Vero cells with swine paramyxoviruses at a MOI of 1.0, 0.1, and 0.01. Virus-specific antigens to the swine viruses were probed with anti-BPIV3 polyclonal bovine antibody (NVSL) (1:100 dilution) followed by antibovine FITC-conjugated antibody. g ISU-92. h Texas-81. i Mock-infected Vero cells. j Optiprep-purified virions of Texas-81 (Lane 2) and ISU-92 (Lane 3) were used for immunoblotting with molecular weight marker (Lane 1). The viral proteins were probed with anti-BPIV3 polyclonal bovine antibody (NVSL), followed by HRP-labeled antibovine IgG (KPL) secondary antibodies
Antigenic analysis
Antigenic analysis at the National Veterinary Services Laboratory (NVSL), Ames, IA indicated that they were closely related to human parainfluenza virus (HPIV) type 1 and 3 and BPIV3 (Table 1). Cross-reactivity in several epitopes in F and HN proteins with hPIV3 and bPIV3 has been reported earlier [34]. Bovine anti-BPIV3 serum was able to detect the cells infected by these viruses (Fig. 2) by IFA. Besides, using bovine anti-BPIV3 serum, all the viral proteins were detectable in OptiPrep-purified virion preparations (Fig. 2). The protein designations described were based on studies with virion proteins of PIV3 [35, 36]. Homologous sera could neutralize virus infectivity of Texas-81 and ISU-92 viruses to similar titers and heterologous serum (BPIV-3-SF) neutralized virus infectivity of Texas 81 at 1:32 and ISU-92 at 1:8, suggesting antigenic variation. The R values of Texas-81 and ISU-92 strains with BPIV3-SF strain were, , respectively, 100 and 35% detected by IFA. These observations suggest that Texas-81 and ISU-92 were antigenically closely related to BPIV3. However, antigenic analysis by IFA also suggested that ISU-92 is a minor subtype of BPIV3, according to the criteria of Brooksby [31].
Transcriptional start and stop sequences
The nucleotide sequence of F and HN genes of both Texas-81 and ISU-92 viruses were determined from RT-PCR products amplified from viral genomic RNA. Individual genes are transcribed by a start–stop mechanism controlled by conserved sequences at the gene borders in members of the family Paramyxoviridae. The F and HN genes of Texas-81 and ISU-92 shared the universal pattern of paramyxovirus genome structure, flanked at 3′ end with a conserved gene-start (GS) sequence and at 5′ end with a gene-end (GE) sequence with an intergenic sequence (IGS) in between. The GS and GE sequences of the F and HN genes were essentially similar to BPIV3. Only the variable fifth and sixth positions of the GS sequence showed host specificity (Table 2). All of them terminated with U-rich GE sequences. The trinucleotide IGS (3′-GAA) of these two viruses were also identical to members of the genera Respirovirus, Morbillivirus, and Henipavirus [37]. A long U-rich sequence was identified in the 3′ untranslated region (UTR) of F gene of ISU-92. This is a characteristic feature commonly found in BPIV3 and HPIV3. This has been shown to result in read-through transcripts of M gene in BPIV3 and HPIV3 [38, 39].
F gene
The F gene of Texas-81 strain was 1,869 nucleotides (nt) in length with a single ORF of 1,620 nt beginning at position 211 (Table 3), capable of encoding a 540-amino acid protein. The F gene of ISU-92 strain was 1,893 nt in length with a 1,620 nt ORF but with a longer 3′ UTR (235 nt). The longer 3′ UTR in ISU-92 strain had a 24-nt “U”-rich insertion compared with the Texas-81 strain. The F protein of ISU-92 had a predicted molecular weight of 60,039 Da and an estimated isoelectric point (pI) of 6.797. For Texas-81 strain, the uncleaved F0 protein had a predicted molecular weight of 60,189 Da and an estimated pI of 6.539. The size determination by immunoblotting confirmed this (Fig. 2). The Texas-81 strain had 100% identity with BPIV3-SF strain both in nucleotide and deduced amino acid sequences. ISU-92 strain had 98.7% identity in nucleotide sequence and 99.1% identity in amino acids with BPIV3-SK217 strain, while it had 90.9 and 95% identity with BPIV3-SF strain in nucleotide and amino acid sequences, respectively (Table 4), for F gene.
The F protein mediates fusion of virus and cell membrane in paramyxoviruses. Fusion activation is dependent on the cleavage of F0 protein into disulfide-linked subunits F2–s–s–F1. The furin, a subtilisin-like endoprotease, is believed to be one of the proteases that cleaves most F proteins intracellularly [40, 41]. In Texas-81 and ISU-92 viruses, the F cleavage motif L/SRTKR was located between amino acid residues 105 and 109, and cleavage occurs between residues 109(R) and 110(F). This cleavage site conformed to the pattern of consensus motif for cleavage by furin [42], R-X-K/R-R, which is conserved in the majority of Paramyxovirinae [32]. The predicted F cleavage site was immediately followed by a 25-amino acid hydrophobic fusion peptide, highly conserved in all paramyxovirus F proteins [32, 43]. The cleaved F1 protein was approximately 51 kDa in size. The F protein of both strains, like the F protein of other paramyxoviruses, was predicted to be a type I membrane protein. The transmembrane region was near the carboxyl terminal (amino acid residues 497–517), which was thought to serve as an anchor in the viral envelope, leaving a 23-amino acid cytoplasmic tail (Fig. 3).
Schematic of the predicted domain structure of the swine paramyxovirus F0 protein and identification of conserved block in F1 (CBF1), conserved block in F2 (CBF2), and fusion peptide (FP) [46, 47] regions were obtained from structure-based sequence alignment of the F protein of SPMV with other paramyxoviruses. Domains are indicated as DI to DIII. TM indicates transmembrane domain. HRA, HRB, and HRC indicate heptad repeat regions. Conserved amino acid residues are shaded
The F proteins of swine paramyxoviruses are essentially similar in structure and function to other paramyxoviruses, and especially to BPIV3. Peptides corresponding to the heptad repeat regions A and B (HRA and HRB) from SV5 [44], hRSV [45], and hPIV3 [46] were conserved in the F protein of SPMV. F proteins assembled into stable six helical bundles (6HBs) (the F1 core), and their structure in HPIV3 had been determined. Structure-based sequence alignment of the SPMV F protein sequences with these 6HB fragments revealed only minor differences. The conserved blocks in F1 and F2 (CBF1, CBF2) were also preserved in SPMV as in other paramyxoviruses which strengthens the hypothesis that these conserved blocks had conserved functions, either in the membrane fusion or in the folding and processing of the F protein [47]. Cysteines, which were important for disulfide bond formation and secondary structure, were also identical to BPIV 3 and HPIV3. Among the five potential conserved N-linked glycosylation sites (N101, N238, N359, N446, and N508) in the F protein [39], all but one had the N-X-T motif. The one at position 446 had the N-X-S motif. It should be noted that only one potential glycosylation site (at N101) was located before the F protein cleavage site, and one site was located in the transmembrane domain (N508). The other three were located prior to the transmembrane domain in the F1 protein and were, therefore, likely to be exposed on the surface, as in other members of the genus Respirovirus [32].
HN gene
The HN gene was 1,888 nt in length with a single ORF beginning at position 74 that could encode a 572-amino acid protein (Table 3). Texas-81 strain shared 99.8% identity with BPIV3-SF strain both in nucleotide and predicted amino acid sequences. ISU-92 strain had 98.4% nucleotide sequence identity and 99.1% amino acid sequence identity with BPIV3-SK217 strain. Identity of ISU-92 with BPIV3-SF strain is 91.7% at nucleotide level and 95.8% at amino acid level (Table 4). Comparing with HPIV3, the two swine viruses had 80.1–82.7% identity in the HN gene nucleotide sequences and 76.4–77.6% identity at deduced amino acid sequences.
The deduced amino acid sequence of the HN protein was 572 residues in length. The HN protein of ISU-92 had a predicted molecular weight of 64,652 Da and an estimated pI of 7.343. The HN protein of Texas-81 strain had a predicted molecular weight of 64,624 Da and an estimated pI of 7.720. The active site residues predicted from NDV or HPIV3 HN crystal structure were also conserved in the swine viruses. The major transmembrane region of HN protein was predicted to be from amino acid residues 36–54 of the protein as in other type II membrane glycoproteins. The transmembrane domain at the N-terminal end of the HN protein contained several conserved substitutions among the examined viruses (Fig. 4).
Schematic of the HN protein globular head region domain structure of swine paramyxovirus. a The transmembrane domain, sialic acid site (NRKSCS motif), and the predicted alpha helix regions (Garnier-Robson) of swine paramyxovirus were given as shaded boxes [48]. Disulfide bonds followed those used in a structure-based sequence alignment and the crystal structure of NDV HN protein [48]. b The corresponding position of the active site residues in the deduced amino acid sequence of the SPMV and other paramyxoviruses
The disulfide bonds in the HN protein C190–C214 (A), C256–C269 (C), C355–C469 (D), C463–C473 (E), C535–C544 (F), C159–C571 (W), and C350–C363 (X) were conserved as in NDV HN or HPIV 3 HN crystal structures [48, 49]. Predicted N-glycosylation sites were conserved in HN proteins as in other members of Paramyxovirinae. In swine viruses, potential N-linked glycan sites observed at all predicted sites (N8, N308, N351, N448 and N523) were similar to HPIV3. The counterpart to N351 in NDV HN is N341, which is glycosylated in that molecule. NDV HN contains a N-linked glycosylation site at N481, but in HPIV3 HN, the sequence at this site is N-P-T and in swine viruses, it is N-P-S, the asparagine moiety of which is thus not expected to be glycosylated. Texas-81 strain had an additional (N15) N-linked glycosylation acceptor site in the HN protein, which appears to be host specific. Whether all of the potential N-linked glycan sites are glycosylated awaits future studies. Both Texas-81 and ISU-92 viruses contained the conserved NRKSCS neuraminidase active site motif [50]. Up to date, all analyzed members of Respirovirus and Rubulavirus had this sequence [51].
The sequence analysis showed that Texas-81 and ISU-92 had higher levels of identity with BPIV-3 than HPIV 1 or HPIV3. The 100% identity at nucleotide level between the F protein of Texas-81 and the SF strain of BPIV3 indicated that cross-species transmission might occur among different hosts; however, further studies will be needed to confirm this. On the other hand, ISU-92, which was closely related to the SK217 and 910N strains of BPIV3, possessed host-specific amino acid residues in the F and HN proteins differing from both BPIV3 strains. There were three host-specific amino acid residues (Y100, A437, and V501) in F protein and three in the HN protein (K70, S88 and S387) of ISU-92 strain, compared to BPIV3 and HPIV3 (Figs. 3, 4). BPIV3 strains SK217 and SF/Ka were shown to differ in virulence [33, 52]. This virulence difference had been associated with a change in amino acid 193 in HN protein which had a dramatic effect on syncytium inducing activity, neuraminidase activity, and hemagglutinating activity [33]. ISU-92 had I at this position, unlike SK217 or 910N, but identical with Texas-81 and BPIV3-SF/Ka. Furthermore, ISU-92 strain was more fusogenic than the Texas-81 strain. The importance of these residues in host specificity awaits future reverse genetic studies.
Phylogenetic analysis
Phylogenetic reconstructions of the full-length F and HN genes of these viruses with other members of the Paramyxoviridae were performed by Parsimony analysis. Phylogenetic analysis of Texas-81 and ISU-92 based on the deduced amino acid sequences of the F and HN proteins placed them on the same clade along with BPIV3 (Fig. 5). Recently, it has been reported that there are two genotypes of BPIV 3: genotype A and genotype B [53]. Genotype B strains have never been reported outside Australia [53]. However, two subgenotypes of BPIV3 were discernible in genotype A: one represented by 910N-like viruses and the other represented by Ka and SF viruses. The Texas-81 and SF/Ka strains formed the first genetic group while ISU-92 and 910N strains formed the second genetic group within genotype A. Phylogenetic reconstruction with HN gene also confirmed these groupings (Fig. 5). Antigenic analysis by IFA has also confirmed that there are subtypic differences between BPIV3 and SPMV strains as well. The above facts support the classification of these two swine viruses in the genus Respirovirus, Paramyxovirinae subfamily.
Phylogenetic analysis. Phylogenetic analysis was performed using Parsimony (PAUP 4.01) software with 1,000 bootstrap replicates. a Phylogenetic analysis based on F gene deduced amino acid sequences compared to other Paramyxoviridae family members. b, c Phylogenetic analysis of PIV3 genotypes based on F and HN gene sequences, respectively
The phylogenetic analyses also indicated that these two strains are very likely the host-adapted variant strains transmitted from cattle to pigs. The host-specific sequence elements might contribute to the growth of the virus in heterogenous hosts [37]. Cross-species infection of BPIV3 in a human [54], BPIV3 in sheep, and ovine PIV-3 in cattle [55] have been reported. This happens to be the first report of cross-species infection of BPIV3 in pigs. Several studies using reverse genetic system to recover recombinant PIV3 have been successfully constructed and employed, including HPIV3 and BPIV3 [56, 57]. Similar approaches can be undertaken to determine the importance of the individual or combined effects of host-specific sequence changes in the pathogenesis and host adaptation of BPIV3. Furthermore, this would also help in developing an attenuated phenotype as a vaccine candidate for these viruses.
References
R.A. Lamb, P. Collins, D. Kolakofsky, J.A. Melero, Y. Nagai, M.B.A. Oldstone, C.R. Pringle, B.K. Rima, Family Paramyxoviridae (Elsevier Academic Press, San Diego, 2005)
J. Moreno-Lopez, P. Correa-Giron, A. Martinez, A. Ericsson, Arch. Virol. 91, 221–231 (1986). doi:https://doi.org/10.1007/BF01314282
T. Linne, M. Berg, A.C. Bergvall, B. Hjertner, J. Moreno-Lopez, Vet. Microbiol. 33, 263–273 (1992). doi:https://doi.org/10.1016/0378-1135(92)90054-W
A.W. Philbey, P.D. Kirkland, A.D. Ross, R.J. Davis, A.B. Gleeson, R.J. Love, P.W. Daniels, A.R. Gould, A.D. Hyatt, Emerg. Infect. Dis. 4, 269–271 (1998)
J. Ellis, L. Hassard, E. Clark, J. Harding, G. Allan, P. Willson, J. Strokappe, K. Martin, F. McNeilly, B. Meehan, D. Todd, D. Haines, Can. Vet. J. 39, 44–51 (1998)
B.H. Janke, P.S. Paul, J.G. Landgraf, P.G. Halbur, C.D. Huinker, J. Vet. Diagn. Invest. 13, 428–433 (2001)
E. Heinen, W. Herbst, N. Schmeer, Arch. Virol. 143, 2233–2239 (1998). doi:https://doi.org/10.1007/s007050050454
S. Tong, M. Li, A. Vincent, R.W. Compans, E. Fritsch, R. Beier, C. Klenk, M. Ohuchi, H.D. Klenk, Virology 301, 322–333 (2002). doi:https://doi.org/10.1006/viro.2002.1594
L. Wang, B.H. Harcourt, M. Yu, A. Tamin, P.A. Rota, W.J. Bellini, B.T. Eaton, Microbes Infect. 3, 279–287 (2001). doi:https://doi.org/10.1016/S1286-4579(01)01381-8
K. Murray, P. Selleck, P. Hooper, A. Hyatt, A. Gould, L. Gleeson, H. Westbury, L. Hiley, L. Selvey, B. Rodwell et al., Science 268, 94–97 (1995). doi:https://doi.org/10.1126/science.7701348
H.E. Field, A.C. Breed, J. Shield, R.M. Hedlefs, K. Pittard, B. Pott, P.M. Summers, Aust. Vet. J. 85, 268–270 (2007). doi:https://doi.org/10.1111/j.1751-0813.2007.00170.x
K.B. Chua, W.J. Bellini, P.A. Rota, B.H. Harcourt, A. Tamin, S.K. Lam, T.G. Ksiazek, P.E. Rollin, S.R. Zaki, W. Shieh, C.S. Goldsmith, D.J. Gubler, J.T. Roehrig, B. Eaton, A.R. Gould, J. Olson, H. Field, P. Daniels, A.E. Ling, C.J. Peters, L.J. Anderson, B.W. Mahy, Science 288, 1432–1435 (2000). doi:https://doi.org/10.1126/science.288.5470.1432
M.S. Chadha, J.A. Comer, L. Lowe, P.A. Rota, P.E. Rollin, W.J. Bellini, T.G. Ksiazek, A. Mishra, Emerg. Infect. Dis. 12, 235–240 (2006)
V.P. Hsu, M.J. Hossain, U.D. Parashar, M.M. Ali, T.G. Ksiazek, I. Kuzmin, M. Niezgoda, C. Rupprecht, J. Bresee, R.F. Breiman, Emerg. Infect. Dis. 10, 2082–2087 (2004)
K. Chant, R. Chan, M. Smith, D.E. Dwyer, P. Kirkland, Emerg. Infect. Dis. 4, 273–275 (1998)
K.B. Chua, L.F. Wang, S.K. Lam, G. Crameri, M. Yu, T. Wise, D. Boyle, A.D. Hyatt, B.T. Eaton, Virology 283, 215–229 (2001). doi:https://doi.org/10.1006/viro.2000.0882
T.R. Bowden, D.B. Boyle, Arch. Virol. 150, 2125–2137 (2005). doi:https://doi.org/10.1007/s00705-005-0552-7
K.B. Chua, L.F. Wang, S.K. Lam, B.T. Eaton, Arch. Virol. 147, 1323–1348 (2002). doi:https://doi.org/10.1007/s00705-002-0815-5
N. Karabatsos, International Catalogue of Arboviruses (American Society of Tropical Medicine and Hygiene, San Antonio, 1985)
M.H. Jun, N. Karabatsos, R.H. Johnson, Aust. J. Exp. Biol. Med. Sci. 55, 645–647 (1977). doi:https://doi.org/10.1038/icb.1977.63
P.J. Jack, D.B. Boyle, B.T. Eaton, L.F. Wang, J. Virol. 79, 10690–10700 (2005). doi:https://doi.org/10.1128/JVI.79.16.10690-10700.2005
Z. Li, M. Yu, H. Zhang, D.E. Magoffin, P.J. Jack, A. Hyatt, H.Y. Wang, L.F. Wang, Virology 346, 219–228 (2006). doi:https://doi.org/10.1016/j.virol.2005.10.039
C.F. Basler, A. Garcia-Sastre, P. Palese, Emerg. Infect. Dis. 11, 108–112 (2005)
H. Schomacker, P.L. Collins, A.C. Schmidt, Virology 330, 178–185 (2004). doi:https://doi.org/10.1016/j.virol.2004.09.019
S. Nylund, M. Karlsen, A. Nylund, Virology 373, 137–148 (2008)
H.H. Nollens, J.F. Wellehan, J.T. Saliki, S.L. Caseltine, E.D. Jensen, W. Van Bonn, S. Venn-Watson, Vet. Microbiol. 128, 231–242 (2008)
A. Kohn, P. Fuchs, J. Virol. 3, 539–540 (1969)
M. Potier, L. Mameli, M. Belisle, L. Dallaire, S.B. Melancon, Anal. Biochem. 94, 287–296 (1979). doi:https://doi.org/10.1016/0003-2697(79)90362-2
O. Ferraris, N. Kessler, B. Lina, Antiviral Res. 68, 43–48 (2005). doi:https://doi.org/10.1016/j.antiviral.2005.07.004
I. Archetti, F.L. Horsfall Jr., J. Exp. Med. 92, 441–462 (1950). doi:https://doi.org/10.1084/jem.92.5.441
J.B. Brooksby, Variants and immunity: definitions for serological investigation, in International Symposium on Foot-and-Mouth Disease: Variants and Immunity, Lyon, France, 1967, pp. 1–10
R.A. Robert, G.D. Parks, in Fields Virology, ed. by B.N. Fields, D.M. Knipe, P.M. Howley, D.E. Griffin (Lippincott Williams & Wilkins, Philadelphia, 2007), pp. 1449–1496
M.M. Breker-Klassen, D. Yoo, L.A. Babiuk, Can. J. Vet. Res. 60, 228–236 (1996)
K.J. Coelingh, C.C. Winter, B.R. Murphy, J.M. Rice, P.C. Kimball, R.A. Olmsted, P.L. Collins, J. Virol. 60, 90–96 (1986)
D.G. Storey, K. Dimock, C.Y. Kang, J. Virol. 52, 761–766 (1984)
S.L. Wechsler, D.M. Lambert, M.S. Galinski, M.W. Pons, J. Virol. 54, 661–664 (1985)
J.E. Bailly, J.M. McAuliffe, M.H. Skiadopoulos, P.L. Collins, B.R. Murphy, Virus Genes 20, 173–182 (2000). doi:https://doi.org/10.1023/A:1008130917204
Y. Sakai, S. Suzu, T. Shioda, H. Shibuta, Nucleic Acids Res. 15, 2927–2944 (1987). doi:https://doi.org/10.1093/nar/15.7.2927
S. Suzu, Y. Sakai, T. Shioda, H. Shibuta, Nucleic Acids Res. 15, 2945–2958 (1987). doi:https://doi.org/10.1093/nar/15.7.2945
H.D. Klenk, W. Garten, Trends Microbiol. 2, 39–43 (1994). doi:https://doi.org/10.1016/0966-842X(94)90123-6
D. Ortmann, M. Ohuchi, H. Angliker, E. Shaw, W. Garten, H.D. Klenk, J. Virol. 68, 2772–2776 (1994)
M. Hosaka, M. Nagahama, W.S. Kim, T. Watanabe, K. Hatsuzawa, J. Ikemizu, K. Murakami, K. Nakayama, J. Biol. Chem. 266, 12127–12130 (1991)
C.M. Horvath, R.G. Paterson, M.A. Shaughnessy, R. Wood, R.A. Lamb, J. Virol. 66, 4564–4569 (1992)
K.A. Baker, R.E. Dutch, R.A. Lamb, T.S. Jardetzky, Mol. Cell 3, 309–319 (1999). doi:https://doi.org/10.1016/S1097-2765(00)80458-X
X. Zhao, M. Singh, V.N. Malashkevich, P.S. Kim, Proc. Natl Acad. Sci. USA 97, 14172–14177 (2000). doi:https://doi.org/10.1073/pnas.260499197
H.S. Yin, R.G. Paterson, X. Wen, R.A. Lamb, T.S. Jardetzky, Proc. Natl Acad. Sci. USA 102, 9288–9293 (2005). doi:https://doi.org/10.1073/pnas.0503989102
A.E. Gardner, R.E. Dutch, J. Virol. 81, 8303–8314 (2007). doi:https://doi.org/10.1128/JVI.00366-07
S. Crennell, T. Takimoto, A. Portner, G. Taylor, Nat. Struct. Biol. 7, 1068–1074 (2000). doi:https://doi.org/10.1038/81002
M.C. Lawrence, N.A. Borg, V.A. Streltsov, P.A. Pilling, V.C. Epa, J.N. Varghese, J.L. McKimm-Breschkin, P.M. Colman, J. Mol. Biol. 335, 1343–1357 (2004). doi:https://doi.org/10.1016/j.jmb.2003.11.032
E.D. Jorgensen, P.L. Collins, P.T. Lomedico, Virology 156, 12–24 (1987). doi:https://doi.org/10.1016/0042-6822(87)90431-4
J.P. Langedijk, F.J. Daus, J.T. van Oirschot, J. Virol. 71, 6155–6167 (1997)
H. Shibuta, T. Kanda, A. Hazama, A. Adachi, M. Matumoto, Infect. Immun. 34, 262–267 (1981)
P.F. Horwood, J.L. Gravel, T.J. Mahony, J. Gen. Virol. 89, 1643–1648 (2008). doi:https://doi.org/10.1099/vir.0.2008/000026-0
Z. Ben-Ishai, V. Naftali, A. Avram, S. Yatziv, J. Med. Virol. 6, 165–168 (1980). doi:https://doi.org/10.1002/jmv.1890060209
R.G. Stevenson, D.E. Hore, J. Comp. Pathol. 80, 613–618 (1970). doi:https://doi.org/10.1016/0021-9975(70)90060-5
M.H. Skiadopoulos, T. Tao, S.R. Surman, P.L. Collins, B.R. Murphy, Vaccine 18, 503–510 (1999). doi:https://doi.org/10.1016/S0264-410X(99)00227-3
M.H. Skiadopoulos, A.P. Durbin, J.M. Tatem, S.L. Wu, M. Paschalis, T. Tao, P.L. Collins, B.R. Murphy, J. Virol. 72, 1762–1768 (1998)
Acknowledgment
The authors thank Dr. X. J. Meng, Dr. Laure Deflube, Shobana Raghunath, Thomas Rogers-Cotrone, and Vrushali Chavan for their excellent technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Qiao, D., Janke, B.H. & Elankumaran, S. Molecular characterization of glycoprotein genes and phylogenetic analysis of two swine paramyxoviruses isolated from United States. Virus Genes 39, 53–65 (2009). https://doi.org/10.1007/s11262-009-0353-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-009-0353-2