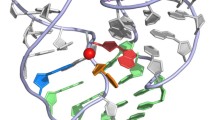
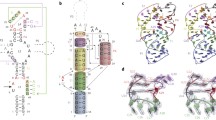
Abstract
Before the discovery of ribozymes, RNA had been proposed to function as a catalyst, based on the discovery that RNA folded into high-ordered structures as protein did. This hypothesis was confirmed in the 1980s, after the discovery of Tetrahymena group I intron and RNase P ribozyme. There have been about ten ribozymes identified during the past thirty years, as well as the fact that ribosomes function as ribozymes. Advances have been made in understanding the structures and functions of ribozymes, with numerous crystal structures resolved in the past years. Here we review the structure-function relationship of both small and large ribozymes, especially the structural basis of their catalysis. ribozyme, structure, catalysis
Similar content being viewed by others
References
Kruger K, Grabowski P J, Zaug A J, et al. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell, 1982, 31(1): 147–157 6297745, 10.1016/0092-8674(82)90414-7, 1:CAS:528:DyaL3sXht1eguw%3D%3D
Guerrier-Takada C, Gardiner K, Marsh T, et al. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell, 1983, 35(3 Pt 2): 849–857 6197186, 10.1016/0092-8674(83)90117-4, 1:CAS:528:DyaL2cXpvFShsA%3D%3D
Butcher S E. Structure and function of the small ribozymes. Curr Opin Struct Biol, 2001, 11(3): 315–320 11406380, 10.1016/S0959-440X(00)00207-4, 1:CAS:528:DC%2BD3MXks1Gqs7Y%3D
Zhou Y, Lu C, Wu Q J, et al. GISSD: Group I intron sequence and structure database. Nucleic Acids Res, 2008, 36(Database issue): D31–37 17942415, 10.1093/nar/gkm766, 1:CAS:528:DC%2BD1cXhtVWitb8%3D
Fedorova O, Zingler N. Group II introns: structure, folding and splicing mechanism. Biol Chem, 2007, 388(7): 665–678 17570818, 10.1515/BC.2007.090, 1:CAS:528:DC%2BD2sXnvVWktLo%3D
Haugen P, Simon D M, Bhattacharya D. The natural history of group I introns. Trends Genet, 2005, 21(2): 111–119 15661357, 10.1016/j.tig.2004.12.007, 1:CAS:528:DC%2BD2MXmslKgtA%3D%3D
Kazantsev A V, Pace N R. Bacterial RNase P: a new view of an ancient enzyme. Nat Rev Microbiol, 2006, 4(10): 729–740 16980936, 10.1038/nrmicro1491, 1:CAS:528:DC%2BD28XpsFOktLk%3D
Cech T R. Structural biology. The ribosome is a ribozyme. Science, 2000, 289(5481): 878–879 10960319, 10.1126/science.289.5481.878, 1:CAS:528:DC%2BD3cXlvFSntLg%3D
Pley H W, Flaherty K M, McKay D B. Three-dimensional structure of a hammerhead ribozyme. Nature, 1994, 372(6501): 68–74 7969422, 10.1038/372068a0, 1:CAS:528:DyaK2MXitVGhtb0%3D
Lilley D M. The origins of RNA catalysis in ribozymes. Trends Biochem Sci, 2003, 28(9): 495–501 13678961, 10.1016/S0968-0004(03)00191-9, 1:CAS:528:DC%2BD3sXnt1ylsLk%3D
Lilley D M. Structure, folding and mechanisms of ribozymes. Curr Opin Struct Biol, 2005, 15(3): 313–323 15919196, 10.1016/j.sbi.2005.05.002, 1:CAS:528:DC%2BD2MXlt1SrsLg%3D
Nakano S, Chadalavada D M, Bevilacqua P C. General acid-base catalysis in the mechanism of a hepatitis delta virus ribozyme. Science, 2000, 287(5457): 1493–1497 10688799, 10.1126/science.287.5457.1493, 1:CAS:528:DC%2BD3cXhsV2qtLw%3D
McKay D B. Structure and function of the hammerhead ribozyme: an unfinished story. RNA, 1996, 2(5): 395–403 8665407, 1:CAS:528:DyaK28XjsFKitbo%3D
Martick M, Scott W G. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell, 2006, 126(2): 309–320 16859740, 10.1016/j.cell.2006.06.036, 1:CAS:528:DC%2BD28Xot1OntL4%3D
Nelson J A, Uhlenbeck O C. When to believe what you see. Mol Cell, 2006, 23(4): 447–450 16916633, 10.1016/j.molcel.2006.08.001, 1:CAS:528:DC%2BD28Xpt1Sntb8%3D
Walter N G, Burke J M. The hairpin ribozyme: structure, assembly and catalysis. Curr Opin Chem Biol, 1998, 2(1): 24–30 9667918, 10.1016/S1367-5931(98)80032-X, 1:CAS:528:DyaK1cXitFygsL0%3D
Rupert P B, Ferre-D’Amare A R. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature, 2001, 410(6830): 780–786 11298439, 10.1038/35071009, 1:CAS:528:DC%2BD3MXjtVemurw%3D
Lafontaine D A, Norman D G, Lilley D M. Structure, folding and activity of the VS ribozyme: importance of the 2-3-6 helical junction. EMBO J, 2001, 20(6): 1415–1424 11250907, 10.1093/emboj/20.6.1415, 1:CAS:528:DC%2BD3MXisVCktrw%3D
Andersen A A, Collins R A. Rearrangement of a stable RNA secondary structure during VS ribozyme catalysis. Mol Cell, 2000, 5(3): 469–478 10882132, 10.1016/S1097-2765(00)80441-4, 1:CAS:528:DC%2BD3cXisVWrsrY%3D
Hiley S L, Sood V D, Fan J, et al. 4-thio-U cross-linking identifies the active site of the VS ribozyme. Embo J, 2002, 21(17): 4691–4698 12198171, 10.1093/emboj/cdf462, 1:CAS:528:DC%2BD38Xms1Kmsrk%3D
Hoffmann B, Mitchell G T, Gendron P, et al. NMR structure of the active conformation of the Varkud satellite ribozyme cleavage site. Proc Natl Acad Sci USA, 2003, 100(12): 7003–7008 12782785, 10.1073/pnas.0832440100, 1:CAS:528:DC%2BD3sXkslOntLc%3D
Perrotta A T, Been M D. A pseudoknot-like structure required for efficient self-cleavage of hepatitis delta virus RNA. Nature, 1991, 350(6317): 434–436 2011192, 10.1038/350434a0, 1:CAS:528:DyaK3MXitVWru7s%3D
Ferre-D’Amare A R, Zhou K, Doudna J A. Crystal structure of a hepatitis delta virus ribozyme. Nature, 1998, 395(6702): 567–574 9783582, 10.1038/26912, 1:CAS:528:DyaK1cXms1aks74%3D
Das S R, Piccirilli J A. General acid catalysis by the hepatitis delta virus ribozyme. Nat Chem Biol, 2005, 1(1): 45–52 16407993, 10.1038/nchembio703, 1:CAS:528:DC%2BD2MXls1Omsrc%3D
Ke A, Zhou K, Ding F, et al. A conformational switch controls hepatitis delta virus ribozyme catalysis. Nature, 2004, 429(6988): 201–205 15141216, 10.1038/nature02522, 1:CAS:528:DC%2BD2cXjvVKgsbw%3D
Xiao S, Scott F, Fierke C A, et al. Eukaryotic ribonuclease P: a plurality of ribonucleoprotein enzymes. Annu Rev Biochem, 2002, 71: 165–189 12045094, 10.1146/annurev.biochem.71.110601.135352, 1:CAS:528:DC%2BD38Xos1ClsbY%3D
Harris M E, Christian E L. Recent insights into the structure and function of the ribonucleoprotein enzyme ribonuclease P. Curr Opin Struct Biol, 2003, 13(3): 325–333 12831883, 10.1016/S0959-440X(03)00069-1, 1:CAS:528:DC%2BD3sXkvVKnsrg%3D
Kazantsev A V, Krivenko A A, Harrington D J, et al. Crystal structure of a bacterial ribonuclease P RNA. Proc Natl Acad Sci USA, 2005, 102(38): 13392–13397 16157868, 10.1073/pnas.0506662102, 1:CAS:528:DC%2BD2MXhtVygsbnK
Torres-Larios A, Swinger K K, Krasilnikov A S, et al. Crystal structure of the RNA component of bacterial ribonuclease P. Nature, 2005, 437(7058): 584–587 16113684, 10.1038/nature04074, 1:CAS:528:DC%2BD2MXhtVajs7vE
Brannvall M, Kikovska E, Kirsebom L. Cross talk between the +73/294 interaction and the cleavage site in RNase P RNA mediated cleavage. Nucleic Acids Res, 2004, 32: 5418–5429 15477392, 10.1093/nar/gkh883
Busch S, Kirsebom L, Notbohm H, et al. Differential role of the intermolecular base-pairs G292-C(75) and G293-C(74) in the reaction catalyzed by Escherichia coli RNase P RNA. J Mol Biol, 2000, 299: 941–951 10843849, 10.1006/jmbi.2000.3789, 1:CAS:528:DC%2BD3cXjvVSku7Y%3D
Evans D, Marquez S M, Pace N R. RNase P: interface of the RNA and protein worlds. Trends Biochem Sci, 2006, 31(6): 333–341 16679018, 10.1016/j.tibs.2006.04.007, 1:CAS:528:DC%2BD28XlvFyrsbk%3D
Cech T R. Self-splicing of group I introns. Annu Rev Biochem, 1990, 59: 543–568 2197983, 10.1146/annurev.bi.59.070190.002551, 1:CAS:528:DyaK3cXltFSjsbY%3D
Doherty E A, Doudna J A. Ribozyme structures and mechanisms. Annu Rev Biochem, 2000, 69: 597–615 10966470, 10.1146/annurev.biochem.69.1.597, 1:CAS:528:DC%2BD3cXnt1ajur4%3D
Scott W G. Ribozymes. Curr Opin Struct Biol, 2007, 17(3): 280–286 17572081, 10.1016/j.sbi.2007.05.003, 1:CAS:528:DC%2BD2sXnsFKlu7k%3D
Shan S, Yoshida A, Sun S, et al. Three metal ions at the active site of the Tetrahymena group I ribozyme. Proc Natl Acad Sci USA, 1999, 96: 12299–12304 10535916, 10.1073/pnas.96.22.12299, 1:CAS:528:DyaK1MXnt1yrt7k%3D
Strobel S, Ortoleva-Donnelly L. A hydrogen-bonding triad stabilizes the chemical transition state of a group I ribozyme. Chem Biol, 1999, 6: 153–165 10074469, 10.1016/S1074-5521(99)89007-3, 1:CAS:528:DyaK1MXhs1yqs78%3D
Michel F, Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol, 1990, 216(3): 585–610 2258934, 10.1016/0022-2836(90)90386-Z, 1:CAS:528:DyaK3MXps1agtw%3D%3D
Adams P L, Stahley M R, Kosek A B, et al. Crystal structure of a self-splicing group I intron with both exons. Nature, 2004, 430(6995): 45–50 15175762, 10.1038/nature02642, 1:CAS:528:DC%2BD2cXlt1CqtL8%3D
Cate J H, Gooding A R, Podell E, et al. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science, 1996, 273(5282): 1678–1685 8781224, 10.1126/science.273.5282.1678, 1:CAS:528:DyaK28XlslGqu7s%3D
Golden B L, Gooding A R, Podell E R, et al. A preorganized active site in the crystal structure of the Tetrahymena ribozyme. Science, 1998, 282(5387): 259–264 9841391, 10.1126/science.282.5387.259, 1:CAS:528:DyaK1cXmsF2jtLs%3D
Golden B L, Kim H, Chase E. Crystal structure of a phage Twort group I ribozyme-product complex. Nat Struct Mol Biol, 2005, 12(1): 82–89 15580277, 10.1038/nsmb868, 1:CAS:528:DC%2BD2MXhtVWltr8%3D
Engelhardt M A, Doherty E A, Knitt D S, et al. The P5abc peripheral element facilitates preorganization of the tetrahymena group I ribozyme for catalysis. Biochemistry, 2000, 39(10): 2639–2651 10704214, 10.1021/bi992313g, 1:CAS:528:DC%2BD3cXhtFWmtrg%3D
Johnson T H, Tijerina P, Chadee A B, et al. Structural specificity conferred by a group I RNA peripheral element. Proc Natl Acad Sci USA, 2005, 102(29): 10176–10181 16009943, 10.1073/pnas.0501498102, 1:CAS:528:DC%2BD2MXmvVelu78%3D
Xiao M, Li T, Yuan X, et al. A peripheral element assembles the compact core structure essential for group I intron self-splicing. Nucleic Acids Res, 2005, 33(14): 4602–4611 16100381, 10.1093/nar/gki770, 1:CAS:528:DC%2BD2MXovVKqtL0%3D
Pichler A, Schroeder R. Folding problems of the 5′ splice site containing the P1 stem of the group I thymidylate synthase intron: substrate binding inhibition in vitro and mis-splicing in vivo. J Biol Chem, 2002, 277(20): 17987–17993 11867626, 10.1074/jbc.M111798200, 1:CAS:528:DC%2BD38XktVCnur4%3D
Szewczak A A, Ortoleva-Donnelly L, Ryder S P, et al. A minor groove RNA triple helix within the catalytic core of a group I intron. Nat Struct Biol, 1998, 5(12): 1037–1042 9846872, 10.1038/4146, 1:CAS:528:DyaK1cXnvFOlu78%3D
Burke J M, Esherick J S, Burfeind W R, et al. A 3′ splice site-binding sequence in the catalytic core of a group I intron. Nature, 1990, 344(6261): 80–82 2406615, 10.1038/344080a0, 1:CAS:528:DyaK3cXhsFaksbo%3D
Chowrira B M, Berzal-Herranz A, Burke J M. Novel system for analysis of group I 3′ splice site reactions based on functional trans-interaction of the P1/P10 reaction helix with the ribozyme’s catalytic core. Nucleic Acids Res, 1995, 23(5): 849–855 7708502, 10.1093/nar/23.5.849, 1:CAS:528:DyaK2MXkvVOltLY%3D
Guo F, Gooding A R, Cech T R. Structure of the Tetrahymena ribozyme: base triple sandwich and metal ion at the active site. Mol Cell, 2004, 16(3): 351–362 15525509, 1:CAS:528:DC%2BD2cXhtVWhtr7E
Zhaxybayeva O, Gogarten J P. Spliceosomal introns: new insights into their evolution. Curr Biol, 2003, 13(19): R764–766 14521854, 10.1016/j.cub.2003.09.017, 1:CAS:528:DC%2BD3sXnvVOmtro%3D
Griffin E A Jr, Qin Z, Michels W J Jr, et al. Group II intron ribozymes that cleave DNA and RNA linkages with similar efficiency, and lack contacts with substrate 2′-hydroxyl groups. Chem Biol, 1995, 2(11): 761–770 9383483, 10.1016/1074-5521(95)90104-3, 1:CAS:528:DyaK2MXps1KnsLk%3D
Qin P Z, Pyle A M. The architectural organization and mechanistic function of group II intron structural elements. Curr Opin Struct Biol, 1998, 8(3): 301–308 9666325, 10.1016/S0959-440X(98)80062-6, 1:CAS:528:DyaK1cXks1eguro%3D
de Lencastre A, Pyle A M. Three essential and conserved regions of the group II intron are proximal to the 5′-splice site. RNA, 2008, 14(1): 11–24 18039742, 10.1261/rna.774008
Wank H, SanFilippo J, Singh R N, et al. A reverse transcriptase/maturase promotes splicing by binding at its own coding segment in a group II intron RNA. Mol Cell, 1999, 4(2): 239–250 10488339, 10.1016/S1097-2765(00)80371-8, 1:CAS:528:DyaK1MXmtVajs7o%3D
Schmidt U, Podar M, Stahl U, et al. Mutations of the two-nucleotide bulge of D5 of a group II intron block splicing in vitro and in vivo: phenotypes and suppressor mutations. RNA, 1996, 2(11): 1161–1172 8903346, 1:CAS:528:DyaK28XmslKmsro%3D
Toor N, Keating K S, Taylor S D, et al. Crystal structure of a self-spliced group II intron. Science, 2008, 320(5872): 77–82 18388288, 10.1126/science.1153803, 1:CAS:528:DC%2BD1cXktVKhtL8%3D
Toor N, Robart A R, Christianson J, et al. Self-splicing of a group IIC intron: 5′ exon recognition and alternative 5′ splicing events implicate the stem-loop motif of a transcriptional terminator. Nucleic Acids Res, 2006, 34(22): 6461–6471 17130159, 10.1093/nar/gkl820, 1:CAS:528:DC%2BD28XhtlCqsr3O
Costa M, Michel F, Westhof E. A three-dimensional perspective on exon binding by a group II self-splicing intron. EMBO J, 2000, 19: 12 10.1093/emboj/19.18.5007
Jacquier A, Jacquesson-Breuleux N. Splice site selection and role of the lariat in a group II intron. J Mol Biol, 1991, 219: 14 10.1016/0022-2836(91)90183-7
Boudvillain M, Pyle A M. Defining functional groups, core structural features and inter-domain tertiary contacts essential for group II intron self-splicing: a NAIM analysis. EMBO J, 1998, 17(23): 7091–7104 9843513, 10.1093/emboj/17.23.7091, 1:CAS:528:DyaK1MXivFCmtQ%3D%3D
Costa M, Michel F. Frequent use of the same tertiary motif by self-folding RNAs. EMBO J, 1995, 14(6): 1276–1285 7720718, 1:CAS:528:DyaK2MXlsFSns7g%3D
Hamill S, Pyle A M. The receptor for branch-site docking within a group II intron active site. Mol Cell, 2006, 23(6): 831–840 16973435, 10.1016/j.molcel.2006.07.017, 1:CAS:528:DC%2BD28XhtVKgu7%2FE
Abramovitz D L, Friedman R A, Pyle A M. Catalytic role of 2′-hydroxyl groups within a group II intron active site. Science, 1996, 271(5254): 1410–1413 8596912, 10.1126/science.271.5254.1410, 1:CAS:528:DyaK28XhsFegtb0%3D
Konforti B B, Abramovitz D L, Duarte C M, et al. Ribozyme catalysis from the major groove of group II intron domain 5. Mol Cell, 1998, 1(3): 433–441 9660927, 10.1016/S1097-2765(00)80043-X, 1:CAS:528:DyaK1cXhtlyhur0%3D
Ban N, Nissen P, Hansen J, et al. The complete atomic structure of the large ribosomal subunit at 2.4 Å, resolution. Science, 2000, 289(5481): 905–920 10937989, 10.1126/science.289.5481.905, 1:STN:280:DC%2BD3cvgslCgsA%3D%3D
Nissen P, Hansen J, Ban N, et al. The structural basis of ribosome activity in peptide bond synthesis. Science, 2000, 289(5481): 920–930 10937990, 10.1126/science.289.5481.920, 1:CAS:528:DC%2BD3cXlvFSnurs%3D
Noller H F. RNA structure: reading the ribosome. Science, 2005, 309(5740): 1508–1514 16141058, 10.1126/science.1111771, 1:CAS:528:DC%2BD2MXpsFWisro%3D
Schuwirth B S, Borovinskaya M A, Hau C W, et al. Structures of the bacterial ribosome at 3.5 Å resolution. Science, 2005, 310(5749): 827–834 16272117, 10.1126/science.1117230, 1:CAS:528:DC%2BD2MXhtFKksLfE
Korostelev A, Trakhanov S, Laurberg M, et al. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell, 2006, 126(6): 1065–1077 16962654, 10.1016/j.cell.2006.08.032, 1:CAS:528:DC%2BD28XhtVCnsrrI
Selmer M, Dunham C M, Murphy F V, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science, 2006, 313(5795): 1935–1942 16959973, 10.1126/science.1131127, 1:CAS:528:DC%2BD28XhtVSnsrbO
Dai L, Zimmerly S. ORF-less and reverse-transcriptase-encoding group II introns in archaebacteria, with a pattern of homing into related group II intron ORFs. RNA, 2003, 9(1): 14–19 12554871, 10.1261/rna.2126203, 1:CAS:528:DC%2BD3sXhtFahsrY%3D
Lewin A S, Hauswirth W W. Ribozyme gene therapy: applications for molecular medicine. Trends Mol Med, 2001, 7(5): 221–228 11325634, 10.1016/S1471-4914(01)01965-7, 1:CAS:528:DC%2BD3MXjvFygurY%3D
Welch P J, Yei S, Barber J R. Ribozyme gene therapy for hepatitis C virus infection. Clin Diagn Virol, 1998, 10(2–3): 163–171 9741642, 10.1016/S0928-0197(98)00029-4, 1:STN:280:DyaK1cvht1KksQ%3D%3D
Duarte E A, Leavitt M C, Yamada O, et al. Hairpin ribozyme gene therapy for AIDS. Methods Mol Biol, 1997, 74: 459–468 9204461, 1:CAS:528:DyaK2sXktVWls78%3D
Watanabe T, Sullenger B A. Induction of wild-type p53 activity in human cancer cells by ribozymes that repair mutant p53 transcripts. Proc Natl Acad Sci USA, 2000, 97: 8490–8494 10890910, 10.1073/pnas.150104097, 1:CAS:528:DC%2BD3cXlt1Ggu7k%3D
Author information
Authors and Affiliations
Corresponding author
Additional information
Supported by National Natural Science Foundation of China (Grant No. 30330170) and National Key Basic Research and Development Program of China (Grant No. 2005CB724604)