Abstract
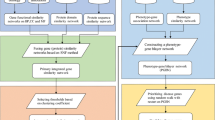
Domains are basic structural and functional unit of proteins, and, thus, exploring associations between protein domains and human inherited diseases will greatly improve our understanding of the pathogenesis of human complex diseases and further benefit the medical prevention, diagnosis and treatment of these diseases. Based on the assumption that deleterious nonsynonymous single nucleotide polymorphisms (nsSNPs) underlying human complex diseases may actually change structures of protein domains, affect functions of corresponding proteins, and finally result in these diseases, we compile a dataset that contains 1174 associations between 433 protein domains and 848 human disease phenotypes. With this dataset, we compare two approaches (guilt-by-association and correlation coefficient) that use a domain-domain interaction network and a phenotype similarity network to prioritize associations between candidate domains and human disease phenotypes. We implement these methods with three distance measures (direct neighbor, shortest path with Gaussian kernel, and diffusion kernel), demonstrate the effectiveness of these methods using three large-scale leave-one-out cross-validation experiments (random control, simulated linkage interval, and whole-genome scan), and evaluate the performance of these methods in terms of three criteria (mean rank ratio, precision, and AUC score). Results show that both methods can effectively prioritize domains that are associated with human diseases at the top of the candidate list, while the correlation coefficient approach can achieve slightly higher performance in most cases. Finally, taking the advantage that the correlation coefficient method does not require known disease-domain associations, we calculate a genome-wide landscape of associations between 4036 protein domains and 5080 human disease phenotypes using this method and offer a freely accessible web interface for this landscape.
Similar content being viewed by others
References
Glazier A M, Nadeau J H, Aitman T J. Finding genes that underlie complex traits. Science, 2002, 298(5602): 2345–2349
Bird T D. Genetic factors in Alzheimer’s disease. The New England Journal of Medicine, 2005, 352(9): 862–864
Lander E S, Schork N J. Genetic dissection of complex traits. Science, 1994, 265(5181): 2037–2048
Wu X, Jiang R, Zhang M Q, Li S. Network-based global inference of human disease genes. Molecular Systems Biology, 2008, 4: 189
Goh K, Cusick M E, Valle D, Childs B, Vidal M, Barabási A L. The human disease network. Proceedings of the National Academy of Sciences of the United States of America, 2007, 104(21): 8685–8690
Domazet-Loso T, Tautz D. An ancient evolutionary origin of genes associated with human genetic diseases. Molecular Biology and Evolution, 2008, 25(12): 2699–2707
Gohlke J M, Thomas R, Zhang Y, Rosenstein M C, Davis A P, Murphy C, Becker K G, Mattingly C J, Portier C J. Genetic and environmental pathways to complex diseases. BMC Systems Biology, 2009, 3: 46
Yu W, Clyne M, Khoury M J, Gwinn M. Phenopedia and Genopedia: disease-centered and gene-centered views of the evolving knowledge of human genetic associations. Bioinformatics, 2010, 26(1): 145–146
Ortutay C, Vihinen M. Identification of candidate disease genes by integrating gene ontologies and protein-interaction networks: case study of primary immunodeficiencies. Nucleic Acids Research, 2009, 37(2): 622–628
Wu X, Liu Q, Jiang R. Align human interactome with phenome to identify causative genes and networks underlying disease families. Bioinformatics, 2009, 25(1): 98–104
Ozgür A, Vu T, Erkan G, Radev D R. Identifying gene-disease associations using centrality on a literature mined gene-interaction network. Bioinformatics, 2008, 24(13): i277–i285
Ideker T, Sharan R. Protein networks in disease. Genome Research, 2008, 18(4): 644–652
Feldman I, Rzhetsky A, Vitkup D. Network properties of genes harboring inherited disease mutations. Proceedings of the National Academy of Sciences of the United States of America, 2008, 105(11): 4323–4328
Kann M G. Protein interactions and disease: computational approaches to uncover the etiology of diseases. Briefings in Bioinformatics, 2007, 8(5): 333–346
Björkholm P, Sonnhammer E L. Comparative analysis and unification of domain-domain interaction networks. Bioinformatics, 2009, 25(22): 3020–3025
Adie E A, Adams R R, Evans K L, Porteous D J, Pickard B S. Speeding disease gene discovery by sequence based candidate prioritization. BMC Bioinformatics, 2005, 6: 55
Aerts S, Lambrechts D, Maity S, Van Loo P, Coessens B, De Smet F, Tranchevent L C, De Moor B, Marynen P, Hassan B, Carmeliet P, Moreau Y. Gene prioritization through genomic data fusion. Nature Biotechnology, 2006, 24(5): 537–544
Chen J, Bardes E E, Aronow B J, Jegga A G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Research, 2009, 37(Web Server issue): W305–W311
Köhler S, Bauer S, Horn D, Robinson P N. Walking the interactome for prioritization of candidate disease genes. The American Journal of Human Genetics, 2008, 82(4): 949–958
Sun J, Jia P, Fanous A H, Webb B T, Van Den Oord E J, Chen X, Bukszar J, Kendler K S, Zhao Z. A multi-dimensional evidence-based candidate gene prioritization approach for complex diseasesschizophrenia as a case. Bioinformatics, 2009, 25(19): 2595–2602
Tranchevent L C, Barriot R, Yu S, Van Vooren S, Van Loo P, Coessens B, De Moor B D, Aerts S, Moreau Y. ENDEAVOUR update: a web resource for gene prioritization in multiple species. Nucleic Acids Research, 2008, 36(Web Server issue): W377–W384
Raghavachari B, Tasneem A, Przytycka T M, Jothi R. DOMINE: a database of protein domain interactions. Nucleic Acids Research, 2008, 36(Database issue): D656–D661
Ng S K, Zhang Z, Tan S H, Lin K. InterDom: a database of putative interacting protein domains for validating predicted protein interactions and complexes. Nucleic Acids Research, 2003, 31(1): 251–254
Ng S K, Zhang Z, Tan S H, Radev D R. Integrative approach for computationally inferring protein domain interactions. Bioinformatics, 2003, 19(8): 923–929
Finn R D, Marshall M, Bateman A. iPfam: visualization of proteinprotein interactions in PDB at domain and amino acid resolutions. Bioinformatics, 2005, 21(3): 410–412
Van Driel M A, Bruggeman J, Vriend G, Brunner H G, Leunissen J A. A text-mining analysis of the human phenome. European Journal of Human Genetics, 2006, 14(5): 535–542
Altshuler D, Daly M, Kruglyak L. Guilt by association. Nature Genetics, 2000, 26(2): 135–137
Wang W, Zhang W, Jiang R, Luan Y. An approach to the discovery of associations of protein domains and complex diseases. In: Proceedings of the Seventh Asia Pacific Bioinformatics Conference. 2009, 908
Wang W. Statistical modeling for analysis of biological high-throughput data and its application. Dissertation for the Doctoral Degree. Jinan: Shandong University. 2009, 51–62
Jain E, Bairoch A, Duvaud S, Phan I, Redaschi N, Suzek B E, Martin M J, McGarvey P, Gasteiger E. Infrastructure for the life sciences: design and implementation of the UniProt website. BMC Bioinformatics, 2009, 10: 136
Finn R D, Tate J, Mistry J, Coggill P C, Sammut S J, Hotz H R, Ceric G, Forslund K, Eddy S R, Sonnhammer E L, Bateman A. The Pfam protein families database. Nucleic Acids Research, 2008, 36(Database issue): D281–D288
Stein A, Panjkovich A, Aloy P. 3did Update: domain-domain and peptide-mediated interactions of known 3D structure. Nucleic Acids Research, 2009, 37(Database issue): D300–D304
Stein A, Russell R B, Aloy P. 3did: interacting protein domains of known three-dimensional structure. Nucleic Acids Research, 2005, 33(Database issue): D413–D417
Lee H, Deng M, Sun F, Chen T. An integrated approach to the prediction of domain-domain interactions. BMC Bioinformatics, 2006, 7: 269
Brunner H G, Van Driel M A. From syndrome families to functional genomics. Nature Reviews Genetics, 2004, 5(7): 545–551
Rhead B, Karolchik D, Kuhn R M, Hinrichs A S, Zweig A S, Fujita P A, Diekhans M, Smith K E, Rosenbloom K R, Raney B J, Pohl A, Pheasant M, Meyer L R, Learned K, Hsu F, Hillman-Jackson J, Harte R A, Giardine B, Dreszer T R, Clawson H, Barber G P, Haussler D, Kent W J. The UCSC genome browser database: update 2010. Nucleic Acids Research, 2010, 38(Database issue): D613–D619
Robinson P N, Köhler S, Bauer S, Seelow D, Horn D, Mundlos S. The human phenotype ontology: a tool for annotating and analyzing human hereditary disease. The American Journal of Human Genetics, 2008, 83(5): 610–615
Lussier Y A, Liu Y. Computational approaches to phenotyping: high-throughput phenomics. Proceedings of the American Thoracic Society, 2007, 4(1): 18–25
Oti M, Huynen M A, Brunner H G. The biological coherence of human phenome databases. The American Journal of Human Genetics, 2009, 85(6): 801–808
Rasmussen C E, Williams C K I. Gaussian Processes for Machine Learning. Cambridge: MIT Press, 2006
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Zhang, W., Chen, Y. & Jiang, R. Comparative study of network-based prioritization of protein domains associated with human complex diseases. Front. Electr. Electron. Eng. China 5, 107–118 (2010). https://doi.org/10.1007/s11460-010-0018-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11460-010-0018-x