Abstract
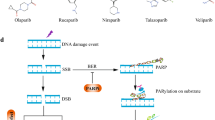
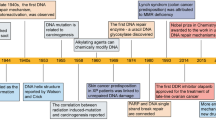
Single-strand break repair protein poly(ADP-ribose) polymerase 1 (PARP1) catalyzes the poly(ADP-ribosyl) ation of many key proteins in vivo and thus plays important roles in multiple DNA damage response pathways, rendering it a promising target in cancer therapy. The tumor-suppressor effects of PARP inhibitors have attracted significant interest for development of novel cancer therapies. However, recent evidence indicated that the underlying mechanism of PARP inhibitors in tumor therapy is more complex than previously expected. The present review will focus on recent progress on the role of PARP1 in the DNA damage response and PARP inhibitors in cancer therapy. The emerging resistance of BRCA-deficient tumors to PARP inhibitors is also briefly discussed from the perspective of DNA damage and repair. These recent research advances will inform the selection of patient populations who can benefit from the PARP inhibitor treatment and development of effective drug combination strategies.
Similar content being viewed by others
References
Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell 2010; 40(2): 179–204
Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med 2009; 361(15): 1475–1485
Friedberg EC, Walker GC, Siede W, Wood RD, Schultz R. DNA Repair and Mutagenesis. USA: Amer Society for Microbiology, 2005
Heitz F, Harter P, Ewald-Riegler N, Papsdorf M, Kommoss S, du Bois A. Poly(ADP-ribosyl)ation polymerases: mechanism and new target of anticancer therapy. Expert Rev Anticancer Ther 2010; 10(7): 1125–1136
Hottiger MO, Hassa PO, Lüscher B, Schüler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci 2010; 35(4): 208–219
Langelier MF, Servent KM, Rogers EE, Pascal JM. A third zincbinding domain of human poly(ADP-ribose) polymerase-1 coordinates DNA-dependent enzyme activation. J Biol Chem 2008; 283(7): 4105–4114
Tao Z, Gao P, Hoffman DW, Liu HW. Domain C of human poly (ADP-ribose) polymerase-1 is important for enzyme activity and contains a novel zinc-ribbon motif. Biochemistry 2008; 47(21): 5804–5813
Hassa PO, Haenni SS, Elser M, Hottiger MO. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol Mol Biol Rev 2006; 70(3): 789–829
de Murcia G, Schreiber V, Molinete M, Saulier B, Poch O, Masson M, Niedergang C, Ménissier de Murcia J. Structure and function of poly(ADP-ribose) polymerase. Mol Cell Biochem 1994; 138(1–2): 15–24
Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer 2010; 10(4): 293–301
Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev 2005; 19(17): 1951–1967
Mangerich A, Bürkle A. How to kill tumor cells with inhibitors of poly(ADP-ribosyl)ation. Int J Cancer 2011; 128(2): 251–265
Martin N, Schwamborn K, Schreiber V, Werner A, Guillier C, Zhang XD, Bischof O, Seeler JS, Dejean A. PARP-1 transcriptional activity is regulated by sumoylation upon heat shock. EMBO J 2009; 28(22): 3534–3548
Casiano CA, Martin SJ, Green DR, Tan EM. Selective cleavage of nuclear autoantigens during CD95 (Fas/APO-1)-mediated T cell apoptosis. J Exp Med 1996; 184(2): 765–770
D’Amours D, Sallmann FR, Dixit VM, Poirier GG. Gain-offunction of poly(ADP-ribose) polymerase-1 upon cleavage by apoptotic proteases: implications for apoptosis. J Cell Sci 2001; 114(Pt 20): 3771–3778
Kaufmann SH, Desnoyers S, Ottaviano Y, Davidson NE, Poirier GG. Specific proteolytic cleavage of poly(ADP-ribose) polymerase: an early marker of chemotherapy-induced apoptosis. Cancer Res 1993; 53(17): 3976–3985
Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet 2008; 9(8): 619–631
Haince JF, Kozlov S, Dawson VL, Dawson TM, Hendzel MJ, Lavin MF, Poirier GG. Ataxia telangiectasia mutated (ATM) signaling network is modulated by a novel poly(ADP-ribose)-dependent pathway in the early response to DNA-damaging agents. J Biol Chem 2007; 282(22): 16441–16453
Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol 2006; 7(7): 517–528
Bryant HE, Petermann E, Schultz N, Jemth AS, Loseva O, Issaeva N, Johansson F, Fernandez S, McGlynn P, Helleday T. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J 2009; 28(17): 2601–2615
Oikawa A, Tohda H, Kanai M, Miwa M, Sugimura T. Inhibitors of poly(adenosine diphosphate ribose) polymerase induce sister chromatid exchanges. Biochem Biophys Res Commun 1980; 97(4): 1311–1316
Wang ZQ, Stingl L, Morrison C, Jantsch M, Los M, Schulze-Osthoff K, Wagner EF. PARP is important for genomic stability but dispensable in apoptosis. Genes Dev 1997; 11(18): 2347–2358
Robinson KM, Schultz MC. Replication-independent assembly of nucleosome arrays in a novel yeast chromatin reconstitution system involves antisilencing factor Asf1p and chromodomain protein Chd1p. Mol Cell Biol 2003; 23(22): 7937–7946
Schultz N, Lopez E, Saleh-Gohari N, Helleday T. Poly(ADP-ribose) polymerase (PARP-1) has a controlling role in homologous recombination. Nucleic Acids Res 2003; 31(17): 4959–4964
Yang YG, Cortes U, Patnaik S, Jasin M, Wang ZQ. Ablation of PARP-1 does not interfere with the repair of DNA double-strand breaks, but compromises the reactivation of stalled replication forks. Oncogene 2004; 23(21): 3872–3882
Citarelli M, Teotia S, Lamb RS. Evolutionary history of the poly (ADP-ribose) polymerase gene family in eukaryotes. BMC Evol Biol 2010; 10(1): 308
Aguilar-Quesada R, Muñoz-Gámez JA, Martín-Oliva D, Peralta A, Valenzuela MT, Matínez-Romero R, Quiles-Pérez R, Menissier-de Murcia J, de Murcia G, Ruiz de Almodóvar M, Oliver FJ. Interaction between ATM and PARP-1 in response to DNA damage and sensitization of ATM deficient cells through PARP inhibition. BMC Mol Biol 2007; 8(1): 29
Hochegger H, Dejsuphong D, Fukushima T, Morrison C, Sonoda E, Schreiber V, Zhao GY, Saberi A, Masutani M, Adachi N, Koyama H, de Murcia G, Takeda S. Parp-1 protects homologous recombination from interference by Ku and Ligase IV in vertebrate cells. EMBO J 2006; 25(6): 1305–1314
Audebert M, Salles B, Calsou P. Effect of double-strand break DNA sequence on the PARP-1 NHEJ pathway. Biochem Biophys Res Commun 2008; 369(3): 982–988
Wang M, Wu W, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res 2006; 34(21): 6170–6182
Mladenov E, Iliakis G. Induction and repair of DNA double strand breaks: the increasing spectrum of non-homologous end joining pathways. Mutat Res 2011; 711(1–2): 61–72
Mansour WY, Rhein T, Dahm-Daphi J. The alternative end-joining pathway for repair of DNA double-strand breaks requires PARP1 but is not dependent upon microhomologies. Nucleic Acids Res 2010; 38(18): 6065–6077
Perrault R, Wang H, Wang M, Rosidi B, Iliakis G. Backup pathways of NHEJ are suppressed by DNA-PK. J Cell Biochem 2004; 92(4): 781–794
Cheng Q, Barboule N, Frit P, Gomez D, Bombarde O, Couderc B, Ren GS, Salles B, Calsou P. Ku counteracts mobilization of PARP1 and MRN in chromatin damaged with DNA double-strand breaks. Nucleic Acids Res 2011; 39(22): 9605–9619
Patel AG, Sarkaria JN, Kaufmann SH. Nonhomologous end joining drives poly(ADP-ribose) polymerase (PARP) inhibitor lethality in homologous recombination-deficient cells. Proc Natl Acad Sci USA 2011; 108(8): 3406–3411
Li B, Navarro S, Kasahara N, Comai L. Identification and biochemical characterization of a Werner’s syndrome protein complex with Ku70/80 and poly(ADP-ribose) polymerase-1. J Biol Chem 2004; 279(14): 13659–13667
Shaheen M, Allen C, Nickoloff JA, Hromas R. Synthetic lethality: exploiting the addiction of cancer to DNA repair. Blood 2011; 117(23): 6074–6082
O’shaughnessy J, Osborne C, Pippen JE, Yoffe M, Patt D, Rocha C, Koo IC, Sherman BM, Bradley C. Iniparib plus chemotherapy in metastatic triple-negative breast cancer. N Engl J Med 2011; 364(3): 205–214
Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005; 434(7035): 913–917
Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005; 434(7035): 917–921
Mendes-Pereira AM, Martin SA, Brough R, McCarthy A, Taylor JR, Kim JS, Waldman T, Lord CJ, Ashworth A. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol Med 2009; 1(6–7): 315–322
Lord CJ, Ashworth A. Targeted therapy for cancer using PARP inhibitors. Curr Opin Pharmacol 2008; 8(4): 363–369
Tong WM, Yang YG, Cao WH, Galendo D, Frappart L, Shen Y, Wang ZQ. Poly(ADP-ribose) polymerase-1 plays a role in suppressing mammary tumourigenesis in mice. Oncogene 2007; 26(26): 3857–3867
Piskunova TS, Yurova MN, Ovsyannikov AI, Semenchenko AV, Zabezhinski MA, Popovich IG, Wang ZQ, Anisimov VN. Deficiency in poly(ADP-ribose) polymerase-1 (PARP-1) accelerates aging and spontaneous carcinogenesis in mice. Curr Gerontol Geriatr Res 2008; 2008: 754190
Morrison C, Smith GC, Stingl L, Jackson SP, Wagner EF, Wang ZQ. Genetic interaction between PARP and DNA-PK in V(D)J recombination and tumorigenesis. Nat Genet 1997; 17(4): 479–482
Tsutsumi M, Masutani M, Nozaki T, Kusuoka O, Tsujiuchi T, Nakagama H, Suzuki H, Konishi Y, Sugimura T. Increased susceptibility of poly(ADP-ribose) polymerase-1 knockout mice to nitrosamine carcinogenicity. Carcinogenesis 2001; 22(1): 1–3
Chiarugi A. A snapshot of chemoresistance to PARP inhibitors. Trends Pharmacol Sci 2012; 33(1): 42–48
Ashworth A. Drug resistance caused by reversion mutation. Cancer Res 2008; 68(24): 10021–10023
Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, Boyd J, Reis-Filho JS, Ashworth A. Resistance to therapy caused by intragenic deletion in BRCA2. Nature 2008; 451(7182): 1111–1115
Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O’Connor MJ, Ashworth A, Carmichael J, Kaye SB, Schellens JH, de Bono JS. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 2009; 361(2): 123–134
Domagala P, Huzarski T, Lubinski J, Gugala K, Domagala W. PARP-1 expression in breast cancer including BRCA1-associated, triple negative and basal-like tumors: possible implications for PARP-1 inhibitor therapy. Breast Cancer Res Treat 2011; 127(3): 861–869
Gottipati P, Vischioni B, Schultz N, Solomons J, Bryant HE, Djureinovic T, Issaeva N, Sleeth K, Sharma RA, Helleday T. Poly (ADP-ribose) polymerase is hyperactivated in homologous recombination-defective cells. Cancer Res 2010; 70(13): 5389–5398
Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, Hiddingh S, Thanasoula M, Kulkarni A, Yang Q, Haffty BG, Tommiska J, Blomqvist C, Drapkin R, Adams DJ, Nevanlinna H, Bartek J, Tarsounas M, Ganesan S, Jonkers J. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol 2010; 17(6): 688–695
Bunting SF, Callén E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, Xu X, Deng CX, Finkel T, Nussenzweig M, Stark JM, Nussenzweig A. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 2010; 141(2): 243–254
Aly A, Ganesan S. BRCA1, PARP, and 53BP1: conditional synthetic lethality and synthetic viability. J Mol Cell Biol 2011; 3(1): 66–74
Wang X, Weaver DT. The ups and downs of DNA repair biomarkers for PARP inhibitor therapies. Am J Cancer Res 2011; 1(3): 301–327
Author information
Authors and Affiliations
Corresponding authors
Additional information
These authors contributed equally to this work.
Rights and permissions
About this article
Cite this article
Wang, Z., Wang, F., Tang, T. et al. The role of PARP1 in the DNA damage response and its application in tumor therapy. Front. Med. 6, 156–164 (2012). https://doi.org/10.1007/s11684-012-0197-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11684-012-0197-3