Abstract
Quantitative real-time PCR (RT-qPCR) is a reliable method for assessing gene expression, provided that suitable reference genes are included to normalize the data. The stability of expression of eight potential reference genes, namely, tubulin (alpha-2,4 tubulin), actin, EF1α (elongation factor 1α), UBC (ubiquitin C), GAPDH (glyceraldehyde-3-phosphate dehydrogenase), psaA (photosynthesis-related plastid gene representing photosystem I), PP2Acs (catalytic subunit of protein phosphatase 2A), and PGK (phosphoglycerate kinase), was assessed in chrysanthemum plants subjected to aphid infestation, heat stress or waterlogging stress using geNorm software. The widely used reference gene EF1α performed well for aphid infested plants but poorly for waterlogged ones. The catalytic subunit of protein phosphatase 2A (PP2Acs) was the best performing one during heat and waterlogging stress, but was the worst during aphid infestation. The commonly used reference gene actin was generally the least stable of the set. No single gene was suitable for normalization on its own. The choice of reference gene(s) is an important factor in gene expression studies based on RT-qPCR.
Similar content being viewed by others
Introduction
The analysis of gene expression provides a useful means of understanding the regulatory networks present in higher organisms [1]. One of the leading methods for achieving this is real time quantitative PCR (RT-qPCR), since this platform is relatively simple to use, has a reasonably broad dynamic range, and a good level of sensitivity [2]. The quantification of gene expression using RT-qPCR requires the simultaneous assay of a reference gene(s), in order to ensure that any variation in signal of the target is not due to variation in the amount of template used to set the PCR. An ideal reference gene is expressed at a level which is largely independent of the experimental conditions to which the subject has been exposed; however, a very few, if any, genes meet this criterion. The choice of an appropriate reference thus requires that they are stably expressed in the actual experimental conditions to be used [3]. Because even this can be difficult to ensure, it has been suggested that a set of reference genes be deployed [4]. The most frequently used reference genes in plant gene expression studies are actin, GAPDH (glyceraldehyde-3-phosphate dehydrogenase), tubulin, and 18s ribosomal RNA [5–7].
Although gene expression has been assessed by RT-qPCR in a wide range of both crop and non-crop plants [6, 8, 9], the technique as yet has not been applied to the important ornamental species chrysanthemum. In this article, we report the stability of eight potential reference genes in chrysanthemum plants exposed to aphid infestation or abiotic (heat or waterlogging) stress.
Materials and Methods
Plant Materials and Treatments
Plants of the chrysanthemum cultivar “Zhongshanzigui” were grown in pots in a greenhouse held at 32°C/27°C, with a light source providing 160 μmol/m2/s photosynthetically active radiation over a 12 h photoperiod. Seedlings at the 6–8 leaf stage were exposed to either a period at 45°C, or infestation with five second instar Macrosiphoniella sanbourni nymphs on each stem tip [10], or a waterlogging treatment consisting of flooding the pots to 2.5 cm above the soil surface for various time periods [11]. The apical shoot of each of three plants was harvested after a 0-, 3-, 6-, 12-, or 24-h exposure to either heat or waterlogging. The aphid-infested plants were similarly sampled in triplicate after 0, 1, 3, 5, and 7 days. After harvesting, the shoots were snap-frozen in liquid nitrogen and stored at −80°C until required.
RNA Extraction
Total RNA was extracted from leaves using the TaKaRa RNAiso reagent, and treated with RNase-free DNaseI (TaKaRa, Japan) [12]. Its concentration was assessed spectrophotometrically at 230, 260 and 280 nm, with samples only being retained where their 260/280 ratio was in the range of 1.80–2.05, and their 260/230 ratio was in the range of 2.00–2.60. The integrity of the RNA was further checked by electrophoresis through 1% agarose gels. The first strand cDNA was synthesized based on 1 μg total RNA using M-MLV reverse transcription system (Promega, USA), according to the manufacturer’s instructions.
Primer Design
The primers directed at the tubulin, UBC, GAPDH, PP2Acs, and PGK sequences (Table 1) were based on chrysanthemum EST sequences [13], which were functionally assigned on the basis of BLAST similarity with homologs from other species. The sequences used for the design of primers targeting actin were obtained from GenBank accessions AB205087. EF1α and psaA were amplified by the degenerate primers (GenBank accessions AB548817 and AB548887, respectively). All the eight primer pairs were designed using Primer Premier v5.0 software (Premier Biosoft International), and their sequences are listed in Table 2.
RT-qPCR
RT-qPCRs were performed in a 25 μl volume, containing 10 μl SYBR Green PCR master mix (TOYOBO, Japan), 10 ng of cDNA template and 0.2 μM of each primer. The cycling conditions comprised an initial denaturation of 95°C/60 s, followed by 40 cycles of 95°C/15 s, 55°C/15 s, and 72°C/45 s. At the end of the cycling process, the temperature was raised from 55 to 95°C at a rate of 0.5°C/s to derive the necessary denaturation curve. Amplicon purity was assumed where a single melting peak was produced. Each qRT-PCR was run in triplicate. PCR efficiency was calculated for each target from the raw fluorescence data taken from Bio-Rad thermal cycler using the LinRegPCR program [14]. Results from the LinRegPCR and Bio-Rad software were imported into Microsoft Excel 2003 and transformed using the comparative Cq method and specific efficiencies for each target [15]. Expression stability was assessed using geNorm software [4].
Results
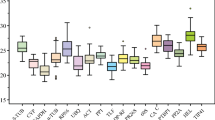
The transcription profiles of the eight reference genes are shown in Fig. 1. The C T values recorded from the plants subjected to stress or aphid infestation were similar to one another. The geNorm software package (http://medgen.ugent.be/genorm) for Microsoft Excel® was applied to identify which of the eight genes would be suitable for normalization purposes. Vandesompele defined the two parameters M (mean expression stability of each individual gene) and V (optimal number of reference genes) using geNorm [4]. Genes associated with a low M values are the most stable, and genes associated with M values >0.15 indicated the need for additional reference gene(s). For aphid infestation, the best performing genes were EF1α and tubulin, having an M value of 0.176. When Used together, the V value was 0.080, indicating no need to add a third reference gene. When the plants were subjected to waterlogging, however, EF1α was no longer stably expressed, and instead PP2Acs with PGK was the most appropriate choice (M = 0.263); curiously, the former gene performed particularly poorly in aphid infested plants (Fig. 2a, c). The most stable gene combination for heat stress was PP2Acs and GAPDH (M = 0.276). Across the entire data set, psaA, EF1α, UBC, and PGK were the most stably expressed genes (Fig. 2d). The V value of 0.185 obtained by the pair of PP2Acs and GAPDH was above the cut-off value, implying that at least one more reference gene was needed. Addition of EF1α lowered the V value to 0.118, so that a fourth reference gene was unnecessary. GeNorm analysis indicated that actin was the least reliable reference gene for heat and waterlogging stress experiments (Fig. 2b, c).
Average expression stability values of control genes by geNorm analysis: a aphid infestation; b heat stress exposure; c waterlogging stress exposure; d all stresses combined. The least stable genes are on the left, and the most stable on the right; e determination of number of reference genes for normalization by geNorm analysis. On the left-most side is the pairwise variation when the number of reference genes is increased from two to three (V2/3). Stepwise inclusion of less stable genes generates the next data points. A decrease in the V value indicates a positive effect and means that the added gene should preferably be included for calculation of a reliable normalization factor
Discussion
Reference genes selection have been reported in a number of plants, such as the following: in potato during late blight exposure, cold, and salt treatment [16], in tomato subjected to nitrogen, cold, and light stress [17], in longan tree during somatic embryogenesis [18], in sunflower throughout six differently controlled leaf senescence conditions [19], and in populus across various developmental stages [20]. In this study, we also find that eight candidate reference genes performed differently upon stresses to which chrysanthemum plants were subjected.
The poor performance of actin in chrysanthemum plants subjected to either heat or waterlogging stress, or aphid infestation was surprising though this gene has been much used as a reference in earlier gene expression studies [21–24]. Recently, several studies have also shown that the use of actin for normalization is not reliable in potato [16], in peach [25], and in bovine muscular tissue [26]. This may be that the actin gene product is a major component of eukaryotic cytoplasmic microfilaments, and it participates in cytoplasmic streaming, cell division and the distribution of the plasma membrane proteins [27].
Genes responsible for elongation factors appear to be stably expressed over a wide range of experimental conditions in potato, peach, and rice [7, 16, 25]. In the present experiments, EF1α was stably expressed under aphid infestation and heat stress, but under waterlogging stress, it was one of the least stable of the eight genes tested (Fig. 1c), indicating that it is not a suitable reference gene under waterlogging stress. However, it was interesting to note that EF1α was always stably expressed in cold-stressed potato and tomato [16, 17], cold and heat- treated rice [7], and similarly, EF1α was stable in chrysanthemum under heat stress. It is inferred that EF1α may be a suitable control gene under cold and heat stress in spite of species.
GAPDH, which has been quite widely applied as a reference gene [28], was indeed the most stable of the genes tested in chrysanthemum plants subjected to heat stress, but was only moderately stable under conditions of either aphid infestation (the fifth most stable gene of the eight genes) or waterlogging stress (the third most stable gene). This gene is therefore also inadequate as a sole reference gene, as has been pointed out on previous examples, and it may lead to wrong results in specific experimental conditions [7, 28]. It may be that GAPDH not only acted as a component of the glycolytic pathway, but also was involved in other processes such as cell proliferation and carcinogenisis [29]. PGK is collocated with GAPDH and also plays important roles in the glycolytic pathway. PGK and GAPDH are potentially co-regulated [30]. Andersen and coworkers mentioned that geNorm may be insensitive to coregulated reference genes [31]. However in our data, potential co-regulation between them was not significant (Fig. 2).
In abiotic (heat or waterlogging) stress, tubulin displayed variable expression patterns. Surprisingly, tubulin showed highly stable expression under aphid infestation among the eight reference genes. However, in perennial ryegrass [32], forage grass Brachiaria brizantha [33], and flax [34], tubuline showed an unacceptable variable expression, but was found to be an appropriate reference gene in cucumber [35], longan tree [18], and in sunflower [19], whereas PP2Acs performed worst under aphid infestation (Fig. 2a), but well under heat or waterlogging stress (Fig. 2b, c). PsaA was the most stably expressed control genes in all combined, but it was not suitable as the best under three different conditions. All these results suggested that reliability of reference genes is highly specific to a particular experimental condition.
The choice of reference genes is an important one, and our results did validate that it is necessary to select suitable reference gene for each experimental condition. Thellin and Vandesompele have recommended the requirement of at least three reference genes [4, 36]. In this study, we have investigated eight reference genes and recommend the use of at least two of the top-ranked reference genes for normalizing gene expression in chrysanthemum leaves under stresses tested here. To our knowledge, this is the first attempt to identify reference genes for gene expression studies in chrysanthemum leaves.
References
Orsel, M., Krapp, A., & Daniel-Vedele, F. (2002). Analysis of the NRT2 nitrate transporter family in Arabidopsis. Structure and gene expression. Plant Physiology, 129, 886–896.
Bustin, S. (2000). Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. Journal of Molecular Endocrinology, 25, 169–193.
Bustin, S. (2002). Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. Journal of Molecular Endocrinology, 29, 23–39.
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., & Speleman, F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome biology, 3(7).
Czechowski, T., Bari, R., Stitt, M., Scheible, W., & Udvardi, M. (2004). Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. The Plant Journal, 38, 366–379.
Jain, M., Nijhawan, A., Tyagi, A., & Khurana, J. (2006). Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochemical and Biophysical Research Communications, 345, 646–651.
Brunner, A., Yakovlev, I., & Strauss, S. (2004). Validating internal controls for quantitative plant gene expression studies. BMC Plant Biology, 4, 14.
Paolacci, A., Tanzarella, O., Porceddu, E., & Ciaffi, M. (2009). Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Molecular Biology, 10, 11.
Jian, B., Liu, B., Bi, Y., Hou, W., Wu, C., & Han, T. (2008). Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Molecular Biology, 9, 59.
Deng, Y., Chen, S., Lu, A., Chen, F., Tang, F., Guan, Z., et al. (2010). Production and characterisation of the intergeneric hybrids between Dendranthema morifolium and Artemisia vulgaris exhibiting enhanced resistance to chrysanthemum aphid (Macrosiphoniellasanbourni). Planta, 231, 693–703.
Yin, D., Chen, S., Chen, F., Guan, Z., & Fang, W. (2009). Morphological and physiological responses of two chrysanthemum cultivars differing in their tolerance to waterlogging. Environmental and Experimental Botany, 67, 87–93.
Miao, H., Jiang, B., Chen, S., Zhang, S., Chen, F., Fang, W., et al. (2010). Isolation of a gibberellin 20-oxidase cDNA from and characterization of its expression in chrysanthemum. Plant Breeding, 129, 707–714.
Chen, S., Miao, H., Chen, F., Jiang, B., Lu, J., & Fang, W. (2009). Analysis of expressed sequence tags (ESTs) collected from the inflorescence of Chrysanthemum. Plant Molecular Biology Reporter, 27, 503–510.
Ramakers, C., Ruijter, J., Deprez, R., & Moorman, A. (2003). Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neuroscience Letters, 339, 62–66.
Livak, K., & Schmittgen, T. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2 −ΔΔCT method. Methods, 25, 402–408.
Nicot, N., Hausman, J., Hoffmann, L., & Evers, D. (2005). Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. Journal of Experimental Botany, 56, 2907–2914.
Løvdal, T., & Lillo, C. (2009). Reference gene selection for quantitative real-time PCR normalization in tomato subjected to nitrogen, cold, and light stress. Analytical Biochemistry, 387, 238–242.
Lin, Y., & Lai, Z. (2010). Reference gene selection for qPCR analysis during somatic embryogenesis in longan tree. Plant Science, 178, 359–365.
Fernandez, P., Di Rienzo, J. A., Moschen, S., Dosio, G. A. A., Aguirrezábal, L. A. N., Hopp, H. E., Paniego, N., & Heinz, R. A. (2010). Comparison of predictive methods and biological validation for qPCR reference genes in sunflower leaf senescence transcript analysis. Plant cell reports, 1–12 (2010). doi:10.1007/s00299-010-0944-3.
Xu, M., Zhang, B., Su, X., Zhang, S., & Huang, M. (2011). Reference gene selection for quantitative real-time polymerase chain reaction in Populus. Analytical Biochemistry, 408, 337–339.
Bezier, A., Lambert, B., & Baillieul, F. (2002). Study of defense-related gene expression in grapevine leaves and berries infected with Botrytis cinerea. European Journal of Plant Pathology, 108, 111–120.
Langer, K., Ache, P., Geiger, D., Stinzing, A., Arend, M., Wind, C., et al. (2002). Poplar potassium transporters capable of controlling K+ homeostasis and K+-dependent xylogenesis. The Plant Journal, 32, 997–1009.
Reid, K., Olsson, N., Schlosser, J., Peng, F., & Lund, S. (2006). An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biology, 6, 27.
Thomas, C., Meyer, D., Wolff, M., Himber, C., Alioua, M., & Steinmetz, A. (2003). Molecular characterization and spatial expression of the sunflower ABP1 gene. Plant Molecular Biology, 52, 1025–1036.
Tong, Z., Gao, Z., Wang, F., Zhou, J., & Zhang, Z. (2009). Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Molecular Biology, 10, 71.
Pérez, R., Tupac-Yupanqui, I., & Dunner, S. (2008). Evaluation of suitable reference genes for gene expression studies in bovine muscular tissue. BMC Molecular Biology, 9, 79.
Stürzenbaum, S., & Kille, P. (2001). Control genes in quantitative molecular biological techniques: the variability of invariance. Comparative Biochemistry and Physiology Part B, 130, 281–289.
Suzuki, T., Higgins, P., & Crawford, D. (2000). Control selection for RNA quantitation. Biotechniques, 29, 332–337.
McNulty, S., & Toscano, W. (1995). Transcriptional regulation of glyceraldehyde-3-phosphate dehydrogenase by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Biochemical and Biophysical Research Communications, 212, 165–171.
Anderson, L., & Carol, A. (2005). Enzyme co-localization in the pea leaf cytosol: 3-P-glycerate kinase, glyceraldehyde-3-P dehydrogenase, triose-P isomerase and aldolase. Plant Science, 169, 620–628.
Andersen, C., Jensen, J., & Orntoft, T. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Research, 64, 5245–5250.
Martin, R. C., Hollenbeck, V. G., & Dombrowski, J. E. (2008). Evaluation of reference genes for quantitative RT-PCR in Lolium perenne. Crop Science, 48, 1881–1887.
Silveira, D., Alves-Ferreira, M., Guimar es, L. A., da Silva, F. R., & Carneiro, V. T. C. (2009). Selection of reference genes for quantitative real-time PCR expression studies in the apomictic and sexual grass Brachiaria brizantha. BMC Plant Biology, 9, 84.
Huis, R., Hawkins, S., & Neutelings, G. (2010). Selection of reference genes for quantitative gene expression normalization in flax (Linum usitatissimum L.). BMC Plant Biology, 10, 71.
Wan, H., Zhao, Z., Qian, C., Sui, Y., Malik, A. A., & Chen, J. (2010). Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Analytical Biochemistry, 399, 257–261.
Thellin, O., Zorzi, W., Lakaye, B., De Borman, B., Coumans, B., Hennen, G., et al. (1999). Housekeeping genes as internal standards: use and limits. Journal of Biotechnology, 75, 291–295.
Acknowledgments
This study is supported by the National Natural Science Foundation of China (Grant No. 30872064, 31071820, 31071825), the Program for Hi-Tech Research, Jiangsu, China, Grant (No. BE2008307, BE2009317, BE2010303), and the Fundamental Research Funds for the Central Universities (KYJ 200907).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gu, C., Chen, S., Liu, Z. et al. Reference Gene Selection for Quantitative Real-Time PCR in Chrysanthemum Subjected to Biotic and Abiotic Stress. Mol Biotechnol 49, 192–197 (2011). https://doi.org/10.1007/s12033-011-9394-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-011-9394-6