Abstract
The therapeutic potential of induced pluripotent stem cells (iPSCs) is well established. Safety concerns remain, however, and these have driven considerable efforts aimed at avoiding host genome alteration during the reprogramming process. At present, the tools used to generate human iPSCs include (1) DNA-based integrative and non-integrative methods and (2) DNA-free reprogramming technologies, including RNA-based approaches. Because of their combined efficiency and safety characteristics, RNA-based methods have emerged as the most promising tool for future iPSC-based regenerative medicine applications. Here, I will discuss novel recent advances in reprogramming technology, especially those utilizing the Sendai virus (SeV) and synthetic modified mRNA. In the future, these technologies may find utility in iPSC reprogramming for cellular lineage-conversion, and its subsequent use in cell-based therapies.
Similar content being viewed by others
Introduction
Human embryonic stem cells (ESC) can provide a potential source of cells for research, regenerative medicine or tissue bioengineering [1]. However, there are limitations that must be overcome, such as immune rejection and ethical and technical issues surrounding the use of human embryos as an ESC source for use in the clinic [2–4]. ESCs represent the prototypical stem cell: they have unlimited clonogenic and self-renewal capacity, and have the pluripotent potential required to all cellular lineages from a single cell. Thus, ESCs under specific stimuli can progress from a pluripotent state, competent to generate all cellular lineages, to a highly committed state characterized by a severe limitation of their differentiatiative potential [5]. This developmental paradigm can be exploited in vitro to direct human ESCs into all the lineages present in the adult organism [6]. Once this process starts, ESCs differentiative potential decreases concurrently with an acquisition of lineage specification [7].
For many years, this process was considered permanent and irreversible. Two key experiments dispute this. The first, performed almost 6 decades ago in John Gurdon's lab, where whole new frogs were developed from reprogrammed “zygotes” obtained by injecting the nucleus from an adult somatic cell into an enucleated oocyte [8–10]. More recently, this observation was taken further when the first mammal (Dolly the sheep) was cloned from an adult somatic cell by nuclear transfer enucleated egg [11]. This experiment essentially defined a new field of somatic cell reprogramming, and, in 2006, Takahashi and Yamanaka identified and defined a set of transcription factors which were able to reprogram somatic cells into a pluripotent state equivalent to that observed in the nuclear transfer studies. These cells were termed induced pluripotent stem cells (iPSCs) [12]. Cells from several different organisms [13–15] and developmental origin have since been reprogrammed into iPSCs using combinations of transcription factors or/and microRNAs (miRNAs) [16–18].
Since the discovery of iPSCs, the notion that cells could be converted from one particular lineage to another, referred to as trans-differentiation, has gained in strength [19]. Indeed, this was first demonstrated 2 decades ago by Weintraub, who showed that a single transcription factor, MyoD, could convert fibroblasts to myoblasts [20]. In addition to this direct lineage conversion process, where one somatic cell is trans-differentiated into another cell type, recent data have established a further new concept termed “indirect” lineage-conversion, in which partial cellular reprogramming takes the cell to an intermediate “plastic” differentiation state from which progenitor-like cells finally differentiate [21–24]. This procedure has been investigated not only in cells cultured in vitro but also in disease target tissues in situ [25, 26].
All these conceptual breakthroughs are most certainly remarkable, but no less impressive have been the delivery techniques developed and used for such reprogramming [18, 27]. In Yamanaka's original work [12], and in around 75 % of published reports, reprogramming has been achieved using either retroviral or lentiviral DNA vector integration. Genome modification in iPSCs represents a safety and therefore a regulatory obstacle for potential clinical application [28] and has therefore been an area of significant interest characterized by a wide variety of different approaches [27].
The initial generation of integrative viral vectors were followed by novel lentiviral vectors carrying poly-cistronic constructs flanked with loxP sites, which permitted Cre-dependent recombination and in order to minimize the footprint of genomic alterations [29, 30]. This solution still carried the risk of insertional mutagenesis, however, and to avoid this, retroviral and lentiviral transduction has been largely replaced by non-integrative methods. Thus, adenoviruses, which are non-integrating vectors, and remain in an epichromosomal form, have also been developed [31]. Unfortunately, adenoviruses are cleared rapidly in dividing cells and gene expression is often not sufficient for efficient reprogramming. Alternatively, the piggyBac (PB) transposon system has allowed transposase-dependent integration and seamless excision of the reprogramming factors after pluripotency has been achieved in two steps by transient transposase expression. Although integrated piggyBac transposon vectors were designed to be removed without trace from the genome, transposition is not always precise, and sequence alterations have been reported in up to 5 % of the transpositions events [32, 33]. Additional non-integrating vector-based plasmids, episomal DNA, and minicircles have been developed to transiently express the reprogramming factors long enough to induce pluripotency [34–36]. However, the efficiency of these approaches remains low and exogenous DNA maintained in the cell becomes a potential risk for insertional mutagenesis and oncogenic transformation. Protein-based technology avoids these hurdles, although suffers from an extremely low efficiency, and requires either chemical treatment or extended periods of transduction [37, 38].
Perhaps the most promising technology that combines efficacy and safety features for future clinical application is based on RNA. Recent reports achieve reprogramming using Sendai virus-based vectors with a single strand RNA phase without DNA intermediate during transduction [39], and sequential transfection of modified RNAs encoding the reprogramming transcription factors [40]. In the rest of this article, I will discuss the novel advances in human reprogramming and some particularly interesting mouse lineage conversion examples, with a special emphasis upon the use of transgene-free RNA for cell-based therapies.
Transcription Factors, miRNAs, and Reprogramming
Somatic cell reprogramming to iPSCs was first achieved by the expression of four different transcription factors: octamer-binding transcription factor 4 (Oct4), SRY-box- containing gene 2 (Sox2), Kruppel-like factor 4 (Klf4), and myelocytomatosis oncogene (Myc) [12]. Soon after, human fibroblasts were successfully reprogrammed using a different combination of factors that included the Nanog homeobox protein (NANOG) and the RNA-binding protein Lin-28 homologue A (LIN28A) or suppressed the oncogenes c-Myc or Klf4 [41]. Given that pluripotency is a tight transcriptionally controlled state was not surprising that several groups demonstrated that miRNAs had roles in the regulation of stem cell differentiation [17]. In fact, specific miRNA families could enhance or inhibit reprogramming demonstrating a role for these RNA molecules in pluripotency homeostasis [42]. Members of specific pluripotency associated miRNAs families, like the miR-302 family, have been shown to drive the initiation of a pluripotent state [16, 43, 44]. Others, such as miR-372 [43], or a combination of miR-200c, miR-302 s and miR-369swere able to enhance reprogramming in human fibroblasts when used in combination with three of the four standard reprogramming factors (Oct4, Sox2, and Klf4) [45]. On the other hand, negative regulation of set of tissue-specific miRNAs that includes miR-21 [46], miR-29a [46], miR-34 [47], and miR-199a-3p [48] demonstrated a suppressive role during reprogramming. In consequence, it is clear that overexpression or suppression of individual miRNAs have profound effects in iPSCs colony formation efficiency and stability [49].
As can be appreciated above, in order to reprogram somatic cells into iPSCs, many reprogramming protocols have been described, using different combinations and variable sets of transcription factors and miRNAs [27]. However, the choice of gene delivery system is the most critical aspect for the efficient and safe generation of iPSCs for future clinical applications. The delivery methods used so far for reprogramming can be classified in three categories depending on host genome alteration risk: DNA integrative (retrovirus, lentivirus, and transposons), non-integrative DNA-base (adenovirus, standard and episomal plasmids, and minicircles), and those that reprogram through a DNA-free approach (proteins, Sendai virus, and synthetic modified mRNA). In this latter group, we find the RNA-based methods that also accomplish the most promising efficiencies for future clinical application (Fig. 1).
Efficiency versus safety to estimate reprogrammed cells future for clinical applications. Initially, reprogramming factors were delivered using DNA-integrative reprogramming methods. So far, these methods include (1) retrovirus, (2) lentivirus, including (3) Cre-loxP-mediated transgene excisable variants, and (4) transposons. These are followed by non-integrative DNA-based tools such as (5) adenovirus, (6) self-replicating episomal and (7) standard plasmids, and (8) minicircles. Finally, DNA-free approaches in nuclear reprogramming had been developed, such as (9) protein transduction and of particular success are the RNA-based tools like (10) the Sendai virus (SeV) and (11) synthetic modified mRNA (modRNA)
DNA-Integrative Reprogramming Tools
DNA-integrative reprogramming methods reported so far include: (1) retrovirus, (2) lentivirus, including Cre-loxP-mediated transgene excisable variants, and (3) transposons.
Retroviral Reprogramming
Most researchers in the field still use integrative viral methods to reprogram differentiated cells into iPSCs, given that they are powerful gene delivery systems and are easily implemented in most research labs. In fact, the first iPSCs were reprogrammed using retroviral vectors to express each of the reprogramming factors [12]. Retroviral vectors can be efficiently transduced into target cells and randomly integrated into the host genome of dividing cells. The reprogramming efficiency reported with this delivery system was 0.01–0.02 % in human cells [50, 51], which was increased to 0.25 % by the addition of hTert and SV-40 large T antigen to Yamanaka's factors [52].
It should be emphasized that it is difficult to compare reprogramming efficiencies because of factors such as subjective criteria to calculate efficiency, use of different combinations of reprogramming factors, great variation in efficiency of different cellular source, and the use of small molecules to enhance reprogramming efficiencies.
Lentiviral Reprogramming
As retroviruses only infect dividing cells, there has been a shift to use lentiviral delivery systems so that both dividing and non-dividing cells could be infected, which means that it could be applied to a wide variety of cell types and improve reprogramming range and efficiency. Original work from the Thomson lab using Sox2 and Oct4 but replacing Klf4 and c-Myc with Nanog and Lin28A generated iPSCs at an efficiency of 0.02 % [41]. In addition, advanced inducible-lentiviral vector systems using doxycycline as an inducing agent have been used to exert specific control of the expression of the four transcription factors [53, 54]. Although acceptable gene delivery has been achieved using lentiviral vectors, concerns have often been raised regarding the incorporation of multiple copies of proviral sequences into the iPSC genome.
To tackle this concern lentiviral vectors were engineered with loxP sites in such a way that the reprogramming integrated factors could be excised by the transient expression of Cre-recombinase [55]. In this Cre/loxP recombination system, a loxP site is inserted into the 3′ long-term repeat (LTR) of self-inactivating (SIN) lentiviral vectors, which contains the reprogramming factors. The loxP site is duplicated into the 5′LTR during proviral replication, resulting in host genomic integration with a transgene flanked by two loxP sites. Taking this approach, fibroblasts obtained from patients have been reprogrammed at efficiency over 1 % [56]. However, after Cre-mediated recombination, a single loxP site flanked by small portions of the 5′ and 3′ LTRs is left behind as a footprint from every insertion/excision event. If more than one fragment remains in an iPSC's genomic DNA, the remaining loxP sites are a susceptible substrate for genome rearrangement [57, 58]. The presence of potentially unstable exogenous DNA is a definite safety concern if differentiated cells from these iPSCs are to be transplanted into patients.
Transposable Elements
In order to overcome problems associated with genomic alterations after viral vectors delivery of reprogramming factors, methods based on transposon/transposase system have been developed as an alternative [32, 33, 59]. PiggyBac (PB) transposons, the most common transposon used in reprogramming, are mobile genetic elements that in the presence of its transposase can be integrated/excised at chromosomal TTAA sites [60]. Recently, different laboratories have used this PB transposon/transposase system, which can be seamlessly removed following stable genomic integration, to successfully generate iPSCs from human fibroblasts at a rate between 0.02-0.05 % [32, 33]. Alternatively, sleeping beauty (SB) transposon system has also been used for cell reprogramming [61, 62]. Although, lack of SB transposon related sequence in human genome minimize potential cross-mobilization between endogenous and transposon sequences in comparison to PB [63], the use of SB transposons implies that TA dinucleotide used as integration sites are changed to TAG(T/A)CTA after excision [64]. This genome edition characteristic would diminish SB interest for future reprogramming use in clinic. Finally, it should be taken into account that transposon reversible integration/excision approach is complex and time consuming since identification of iPSCs with minimal-copy insertions, mapping of integration sites, excision of the reprogramming cassette, and validation of factor-free clones is required.
Non-Integrative DNA-Based Methodology
One of the major conceptual advances in the development of safer reprogramming technologies for clinical application was the observation that the integration of transcription factors into the genome is not required for the reprogramming of somatic cells. Non-integrative DNA-based tools used include (1) adenovirus, (2) standard and self-replicating episomal plasmids, and (3) minicircles.
Reprogramming with Adenoviral Vectors
Adenoviral vectors are non-integrating double-stranded DNA vectors that remain in epichromosomal form in cells [65]. At first glance, this appears to be an excellent alternative expression vehicle for generating iPSCs. However, the reprogramming efficiency of this method is only around 0.0002 % in human cells. Consequently, the reprogramming efficiency must be significantly improved before this delivery method can become clinically useful.
Standard and Self-Replicating Episomal Plasmids
Ectopic expression of reprogramming factors from an episomal plasmid has also been explored as a possibility to generate footprint-free iPSCs. Unfortunately, transient transfection with a non-autonomous replicating plasmid does not result in expression for a sufficient period of time to reprogram somatic cells efficiently to iPSCs, and sequential transfections are required to achieve iPSCs [66].
Unlike regular plasmids, episomal vectors derived from Epstein-Barr virus that contain oriP/EBNA1 sequences can self-replicate once per cell cycle, albeit inefficiently. For this reason, iPSC lines normally lose the plasmid by their 15th passage in culture. Yu et al. successfully generated the first human iPSCs using an oriP/EBNA1 episomal plasmid containing reprogramming factors, but reprogrammed colonies were observed at a very low frequency (0.0003–0.0006 %) [67]. After clonal analysis, one third of subclones from two of the original iPSC lines had lost the episomal plasmid. Building on these results, Yamanaka and colleagues improved reprogramming efficiencies adding a short hairpin RNA (shRNA) against the tumor suppressor p53 to the cocktail to dedifferentiate adult peripheral blood mononuclear cells [34, 68]. However, p53 knockdown may raise concerns about genomic instability of iPS generated. Recently, this problem has been bypassed using, in the reprogramming mix, the anti-apoptotic factor Bcl-xl instead of the shRNA against p53 [69]. Nevertheless, this technology will need further development to be applied therapeutically.
Non-Viral Minicircles
Minicircle vectors are circularized vectors in which the plasmid backbone has been released leaving only the eukaryotic promoter and cDNA(s) that are to be expressed. Using a minicircle vector expressing Lin28A, Nanog, Sox2,Oct4, and a GFP marker in human adipose stromal cells was able to reprogram 0.005 % of the cells. Surprisingly, this method was even less efficient at reprogramming neonatal fibroblasts, and no other reports have successfully reprogrammed other somatic cells [70, 71]. Therefore, more validation will be required before this method can be widely used.
DNA-Free Approaches
One common drawback of the delivery systems presented above, especially for those who want to take iPSCs from bench to bed, is the use of exogenous DNA sequences which are either integrated into the host genome or have the potential risk to do so. Growing concerns over the presence of transgene sequences in iPSCs have necessitated a number of astonishing recent technical developments. New tools include nonintegrative viral vectors or DNA-free approaches in nuclear reprogramming, such as (1) protein transduction, and in particular, RNA-based tools like (2) Sendai virus (SeV) and (3) synthetic modified mRNA (modRNA).
Reprogramming by Protein Transduction
Direct protein transduction of reprogramming factors allows generation of footprint-free iPSCs. As such, this method could be another good choice for the creation of iPSCs suitable for studies in translational medicine. Unfortunately, it has been technically challenging to synthesize large amounts of bioactive proteins that can cross the plasma membrane, and very low efficiencies of 0.001 % in human cells have been reported [37, 38].
Viral RNA-Based Reprogramming Using Sendai Virus (SeV)
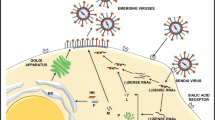
SeV-based vectors are able to induce reprogramming factor expression without entering the nucleus of an infected cell and, crucially, this approach avoids any DNA phase, as the viral genome remains as RNA in the cytoplasm [72]. SeV is an enveloped virus member of the family Paramyxoviridae with a nonsegmented negative-strand RNA genome [73]. The SeV genome has 15,384 nucleotides and includes six cistrons [74] (Fig. 2), which are organized as a single negative-strand RNA molecule. SeV genomic RNA forms a complex with nucleoprotein (NP), phosphoprotein (P), the small subunit of RNA polymerase, and the catalytic subunit of the polymerase called large protein (L) to form the ribonucleoprotein (RNP) macromolecule. Once in the cytoplasm, RNPs act as the template for transcription and replication. Matrix protein (M) engages in the assembly of viral particles with a large spherical shape and average diameter of 260 nm. Two envelope glycoproteins, hemagglutinin-neuraminidase (HN) and fusion (F) protein, which are integrated into lipid bilayers, mediate the attachment of virions to allow RNPs to penetrate into target cells. Viral infection depends on HN protein that recognizes the sialic acid, which is present as a glycoprotein or glycolipid on the cell surface [75]. Sialic acid is widely expressed in mammalian cells, enabling SeV to target a broad cell type range [76–79]. The second envelope component, F protein, is synthesized as an inactive precursor protein F0 and split into F1 and F2 by proteolytic cleavage. Processed F protein penetrates into the cellular membrane to induce the bilipidic layer and viral envelope to merge [80]. Once inside the infected cell, SeV vectors rely only on the virus-encoded RNA polymerase and ubiquitous cellular tubulin for their gene expression [81]. In addition, SeV replication is independent of nuclear factors and does not involve a DNA phase. Therefore, it does not transform cells by integrating its genetic information into the cellular genome.
Sendai virus (SeV) vector structure and recombinant infective particle generation. a Schematic representation of wild-type RNA SeV genomic organization. Infectively competent SeV-based vectors from first generation express genes of interest (GOI) from a 3′ wild-type SeV vector together with viral proteins NP, P, M, F, HN, and L (see text for details). Second-generation SeV vectors (infectively incompetent), include a T7 promoter and carry an F-defective SeV gene and termosensitive replicative proteins L* and P* genes. b Schematic representation of the two-step procedure for recovery of the F defective SeV vector. In the first step, the functional RNPs are recovered in LLC-MK2 or HEK293Tcells by expressing the viral proteins L, P, and NP together with T7 RNA polymerase. In the second step, RNPs are introduced via a cationic liposome to F-expressing LLC-MK2 cells (LLC-MK2/F7) to produce infectious F-defective virions
Since 1995, when the first recombinant SeV reconstitution from full-length genomic cDNA was completed, growing interest pushed forward the development of safer SeV vectors for clinical application [82]. Several SeV vectors expressing genes of interest (GOI) have been generated based on the wild-type SeV strain (Fig. 2). The first generation SeV vectors used the region between the 3′ terminus and the NP gene of a full-length SeV genome to insert the GOI. At this location, SeV vectors maintain the replication capabilities and efficiently produced the GOI protein product when cultured in fertilized chicken eggs [83]. However, for potential medical applications, defective SeV vectors that are unable to fuse cellular membranes were required; as a result of which, SeV vectors in which the F gene was deleted were developed.
These second generation vectors used F-defective SeV and a GOI [72, 84]. In principle, this SeV should be self-replication competent but unable to infect neighbor cells. Therefore, the recombinant SeV virus including a T7 promoter at the 3′ recovery in the laboratory must be achieved in two steps (Fig. 2). The first step involves RNPs generation from LLC-MK2 or HEK293T cells using an F-defective cDNA clone and plasmids expressing NP, P, L (F5R in alternative protocols) and the T7 RNA-polymerase genes. The second step is the isolation and transfection of functional RNPs into the F-expressing packaging cell line (LLC-MK2/F7), followed by collection of infectious particles from the supernatants. This second-generation SeV vector does not encode F protein itself, but instead incorporates it when expressed in trans from the packaging cells. Thus, using this approach in human fibroblasts, transgene-free iPSCs have been generated with efficiencies over 1.5 % [39]. It should be noted that different cell types including fibroblasts, CD34+ cord blood cells [85], and activated T-lymphocytes [86, 87] have been successfully reprogrammed using SeV F protein-deficient and termosensitive (ΔF/TS) vectors to date. Fully reprogrammed iPSCs with Yamanaka factors Oct4, Sox2, Klf, and c-Myc are able to passively eliminate viral RNA through successive cell passages [39]. To achieve more effective RNA viral removal mutations in P and/or L replication, additional genes have been introduced to confer temperature sensitivity and then interfere with the RNP complex stability and viral replication [88]. Importantly, it should be emphasized that improved recombinant SeV for nuclear reprogramming has been refined and are now commercially available.
To date, SeV vectors have proven to be an efficient method to deliver transgenes into a wide range of host cell species and tissues [89]. SeV vectors have been already clinically applied in a gene therapy setting for different diseases such as cystic fibrosis [77, 90], critical limb ischemia [91], and vaccines for AIDS [92, 93]. In the nuclear reprogramming context, the advantages brought by SeV technology are the following: (1) it is nonpathogenic to human, (2) it has a high efficiency of infection in dividing and quiescent cells, (3) it results in high levels of gene expression, (4) it is not integrative, and (5) it is controllably removable. Hence, recombinant SeV vectors are powerful tools for basic research, molecular therapy, and in regenerative medicine [89, 93].
Synthetic Modified mRNA (modRNA) for iPSC Derivation
The ability to express reprogramming factors as mRNA offers another method to make DNA transgene-free iPSCs. Several technical hurdles have had to be overcome before making this a practical possibility, including efficient RNA synthesis, stability, and translation [94] and lack of immunogenicity [95, 96]. Initially, to generate efficiently enough copies of target RNAs, chemical synthesis was unsuitable. The major technical obstacle was the limited yield obtained with this method. Yield decreases exponentially as transcripts grow since coupling efficiencies at each step are between 90–99 %. Thus it was not feasible to synthesize chemically very long RNA molecules. In contrast, bacteriophage polymerases, such as T7 RNA polymerase, are processive, and it is therefore possible to use in vitro transcription (IVT) to generate long RNA transcripts [97]. These long RNA molecules have recently been used as reprogramming tools for the generation of iPSCs [40, 98].
In order to prepare the reprogramming DNA templates for IVT, the substrates include a T7 promoter, 5′ UTR, 3′ UTR, and a poly-(A) tail to stabilize the RNA products [97] (Fig. 3). Given the labile nature of RNA molecules, a key step in cellular mRNA processing is the addition of a 5′ cap structure, where 5′-5′ triphosphate is linked between the 5′ end of the RNA and a guanosine nucleotide. The cap is methylated enzymatically at the N-7 position of the guanosine to form mature mCAP [99]. Processed 5′ cap adds stability to mRNA enhancing as well translation. Consequently, to stabilize modRNAs a cap have been incorporated to transcribed mRNA including a mixture of cap analog and GTP. The first cap analog used during IVT to generate stable modRNAs was mCAP [m7G(5′)ppp(5′)G] (Fig. 3). The inherent nature of the molecule implies that only 50 % of the time, mCAP was inserted in the correct orientation to enhance translation. The other 50 % of molecules were not substrates for efficient translation, reducing the specific activity of the RNA transcript to half. To avoid this problem, anti-reverse cap analog (ARCA) was introduced [3′ O-Me-m7G(5′)ppp(5′)G] [100] (Fig. 3). Therefore, ARCA can only be inserted in the proper orientation, resulting in capped modRNAs that ones in the cell are translated twice as efficiently as those capped with mCAP. Although an improvement, and taking into account that reprogramming factor expression is typically robust only for about 24 h, whilst iPSCs generation requires several weeks to complete, there exists a need for successive transfections. This is an obvious handicap, since mRNA transfection induces the innate immune responses by activation of Toll-like receptors [95] (TLR3, TLR7, and TLR8) and RNA sensors [96, 101] (RIG-I and PKR). Consequently, transfection of mRNAs into mammalian cells results in severe cytotoxicity. Surprisingly, incorporation of certain naturally occurring modified nucleosides into modRNA suppressed the activation of the innate immune response [102]. As a result, 5-ribosyluracil (pseudo-UTP) [98, 103] and 5-methylcytidine-5′-triphosphate (5-Methyl-CTP) substitutions reduce in vitro toxicity. This reduced cytotoxicity has been critical for successful iPSC reprograming: several rounds of modRNA transfection can now be achieved.
DNA template structure and key molecules used during modified mRNA (modRNA) synthesis. Linear in vitro transcription (IVT) templates incorporate a T7 promoter, 5′ and 3′ untranslated regions (UTRs) flanking an insertion site designed to accept a gene of interest (GOI) and a poly (A) tail. Modified capping molecule and nucleotides structure used for synthesis are also shown: anti reverse cap analog (ARCA) 3′-O-Me-m7G(5′)ppp(5′)G, pseudouridine-5'-triphosphate (Pseudo-UTP) and 5-methylcytidine-5′-triphosphate (5-Methyl-CTP)
Pseudouridine (Fig. 3) was the first modified ribonucleoside discovered and the most abundant natural modified RNA base; as such, it has been considered the “fifth RNA nucleoside”. It can be found in structural RNAs, such as transfer, ribosomal, and small nuclear RNAs. On the other hand, 5-Methyl-CTP (Fig. 3) can also be found in RNA molecules such as mRNA, miRNA, and tRNA as methylation of the position 5 of cytidine, given it is a common substrate for post-transcriptional modification [104]. In the context of IVT, complete substitution of cytidine and uridine by pseudo-UTP and 5-Methyl-CTP increase modRNA stability against nuclease activities and translation, and reduce cytotoxicity by avoiding the activation of the immune response [40]. These nucleoside substitutions allow robust and sustained protein expression. Combined with neutralizing type I interferon receptor supplements to completely suppress residual immune response, regular transfection of modRNAs encoding reprogramming factors (Oct4, Sox2, Klf4, c-Myc, and Lin28A) has led to successful iPSC reprogramming with efficiencies over 4 %. ModRNA technology allows iPSCs generation without residual traces of transgenes, making it an attractive option for cell-based therapies in translational research [105]. Indeed, recent reports have demonstrated a potential therapeutical application for modRNA in vivo. Using a lung disease mouse model that lack surfactant protein B expression in the pulmonary epithelium, Kormann and colleagues have been able to rescue the wild-type phenotype by expressing surfactant protein B from a synthesized modRNA [106]. Also mentioned here is that on top of this localized use, two groups have demonstrated that modRNA can be used to produce a systemic effect, over biologically active erythropoietin in vivo [106, 107].
Future Perspective for RNA-Technology in Cell-Fate Derivation
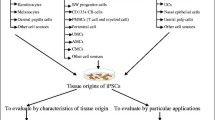
The possibility of generating pluripotent cells from adult individual has obviously created enormous expectations, and has the potential to revolutionize the field of regenerative medicine. Once obtained, iPSCs are just the starting point for autologous cell therapies, and subsequent differentiation is essential. In other words, the final aim for cell-based therapies is to obtain enough committed progenitors or fully differentiated cells at will. In that sense, there are three possible sources from where to obtain these cells: (1) terminal differentiation from iPSCs (pluripotent state) to the required somatic cell types (ground state) [108–111]; (2) indirect lineage conversion from a somatic cell to a dedifferentiated activated state (progenitor-like state) that allows the commitment into several final cell lines in response to environmental signals and/or transcription factors [22, 24, 112, 113]; and (3) somatic cell direct linage conversion or trans-differentiation avoiding a progenitor-like state [23, 25, 26, 114–116] (Fig. 4).
Pathways to generate specific differentiated lineages from specific somatic cells. Fully differentiated target cells can be induced by three conceptually separate mechanisms: reprogramming (1) by ectopic expression of Yamanaka factors to induce a pluripotent state which can be later differentiated towards all lineages, (2) by indirect lineage-conversion where an activation phase is required to generate precursor-like cells, or (3) by direct lineage conversion with forced expression of lineage-specific transcription factors. Epigenetic modifications during indirect lineage-conversion are milder compared with the reprogramming process where all epigenetic marks are erased, in contrast to direct conversion which does not imply epigenetic modification
Forced expression of reprogramming factors induces a global dedifferentiation phenotype, which involves the removal of epigenetic marks and the reestablishment of the pluripotency network [117]. Most differentiation protocols are inefficient, and derivation to several cell types is often complex. Cardiomyocyte differentiation of human iPSCs was first achieved by Zhang et al. who used the spontaneous embryoid-body based differentiation method [109, 110]. Some of the generated myocytes have been demonstrated to display molecular, structural, and functional properties of early human cardiomyocytes, showing different electrophysiological properties with ventricular-like, atrial-like, and nodal-like potential features [108, 110]. Human-derived cardiomyocyte are also able to display functional syncytium with stable pacemaker activity and synchronize action potential propagation [118]. RNA-based tools are the most efficient and safest methods for iPSC generation [40] (Fig. 1), and it is likely that these techniques could contribute greatly to the development of new differentiation procedures. Thus, it could be possible that SeV vectors or modRNA transient transfection encoding differentiation factors could be used to reset the epigenetic marks of a given cell type. Improvements in the differentiation efficiency would have a great impact on bringing iPSC technology closer to clinical application.
Direct lineage conversion, which does not involve an activation state, depends on whether defined factors are able to override epigenetic marks and drive in trans the establishment of the new target cell's genomic identity. Mouse fibroblasts, for instance, have been directly converted into cardiomyocytes by forced expression of cardiac-specific transcription factors Gata4, Mef2c, and Tbx5 alone [119], or together with Hand2 [26]. Furthermore, in vivo trans expression of these genes in mouse heart fibroblasts after myocardial infarction can induce transdifferentiation into functional cardiomyocytes and improve heart function [25, 26]. These examples point to SeV tools as ideal for future use during direct lineage-conversion for several reasons. First, to overcome the existing epigenetic marks, strong and continuous expression of the trans-differentiating factors are crucial in order to induce direct lineage conversion. As discussed above, SeV constructs can drive ectopic overexpression of defined factors in any cell type that expresses tubulin. Second, specific epigenetic marks determine the accessible sites to which the trans-differentiating factors are able to transcribe from. Then, for a given combination of factors, the success in establishing a new determined network typical of a target cell will depend on matching genome accessibility of the original cell type. Hence, finding the best cellular source in combination with specific factors to directly convert one cell type into another could be challenging, although in the cardiac setting, heart-resident fibroblasts appear as the best substrate. As a typical recombinant viral vector, SeV is able to deliver transgenes more efficiently than a nonviral system. Its exceptionally broad host range gives SeV system a significant advantage over other methods. Since direct lineage-conversion occurs in the absence of a pluripotent state and generates post-mitotic populations, it could also theoretically reduce the risk of uncontrolled post-transplantation cell proliferation.
Indirect lineage conversion requires an activation state that leads to the generation of cellular intermediates, in which epigenetic marks get re-written [21]. In this case, activated cells acquire a precursor-like phenotype with multipotent differentiation capacity. This is important, as generation of progenitor cells with such capabilities will expand applications in regenerative medicine, specifically in cases where progenitor transplantation might be an advantage over fully differentiated cells. Recent reports have demonstrated that short temporal expression of pluripotency factors was enough to induce a partially de-differentiated state suitable for conversion into specific cell types by extracellular developmental signals [113]. More striking was the fact that cells in a more pluripotent stage diminished their lineage conversion efficiency, like it has been shown for mouse cardiomyocyte differentiation [119]. These data indicate that the process itself must be fine-tuned in order to achieve partial reprogramming and start a differentiation route on time to obtain the target cell of interest. Synthetic modRNA technology presents a number of characteristics that make it a potential powerful platform for this type of indirect linage conversion. These features include the fact that modRNA enables robust and dose-titrable translation of nearly any protein. Moreover, since modRNA combination of multiple transcripts can be transfected into cells at once, co-translation of several factors at desired stoichiometry is simply controlled by changing the dose of the relevant modRNAs [120]. To our knowledge, no other reprogramming technology permits such control over reprogramming factor expression. Remarkably, the labile nature of modRNAs inside the cells, (its half-life of around 24 h was originally considered a serious handicap) has become a powerful characteristic, differentiating it from alternative reprogramming vectors. As a consequence, modRNA stands out as an ideal tool to temporally and quantitatively control the expression of any given combination of factors in order to redefine cellular fate.
It is also noteworthy that RNA-based reprogramming methodology could easily take advantage of synthetic biology for further technical development. As emphasized throughout this review, using DNA-free delivery techniques abolishes the risk of random genomic integration and opens up the opportunity to develop safe artificial tools for reprogramming and/or lineage conversion. A recent example has demonstrated that reprogramming could be enhanced using engineered variants of Oct4 fused to N-terminal MyoD transactivation domain [121]. It is known that ectopic expression of MyoD is able to direct the fate of iPSCs towards a myogenic fate [122], and is also able to induce trans-differentiation. Hence, this synthetic transcription factor maintains the powerful transactivation activity of MyoD, without losing the target specificity of Oct4. By modRNA transfection of this engineered factor chromatin accessibility and recruitment of chromatin remodeling proteins to the Oct4 site can be increased, resulting in a radical acceleration of iPSCs derivation [105].
Taken together, it seems possible that RNA-based technologies for reprogramming and encoding lineage specification factors could emerge as important tools for generating diverse cell types, either by terminal differentiation from iPSCs, or by direct or indirect lineage conversion, for experimental and future therapeutic applications.
References
Gu, Q., Hao, J., Zhao, X. Y., et al. (2012). Rapid conversion of human ESCs into mouse ESC-like pluripotent state by optimizing culture conditions. Protein & Cell, 3, 71.
Thomson, J. A., Itskovitz-Eldor, J., Shapiro, S. S., et al. (1998). Embryonic stem cell lines derived from human blastocysts. Science, 282, 1145.
Saric, T., Frenzel, L. P., & Hescheler, J. (2008). Immunological barriers to embryonic stem cell-derived therapies. Cells, Tissues, Organs, 188, 78.
Giacomini, M., Baylis, F., & Robert, J. (2007). Banking on it: public policy and the ethics of stem cell research and development. Social Science & Medicine, 65, 1490.
Cho, L. T., Wamaitha, S. E., Tsai, I. J., et al. (2012). Conversion from mouse embryonic to extra-embryonic endoderm stem cells reveals distinct differentiation capacities of pluripotent stem cell states. Development, 139, 2866.
Murry, C. E., & Keller, G. (2008). Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell, 132, 661.
Yu, J., & Thomson, J. A. (2008). Pluripotent stem cell lines. Genes & Development, 22, 1987.
Gurdon, J. B. (1968). Transplanted nuclei and cell differentiation. Scientific American, 219, 24.
Gurdon, J. B. (1968). Changes in somatic cell nuclei inserted into growing and maturing amphibian oocytes. Journal of Embryology and Experimental Morphology, 20, 401.
Gurdon, J. B., Elsdale, T. R., & Fischberg, M. (1958). Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature, 182, 64.
Campbell, K. H., McWhir, J., Ritchie, W. A., & Wilmut, I. (1996). Sheep cloned by nuclear transfer from a cultured cell line. Nature, 380, 64.
Takahashi, K., & Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell, 126, 663.
Liao, J., Cui, C., Chen, S., et al. (2009). Generation of induced pluripotent stem cell lines from adult rat cells. Cell Stem Cell, 4, 11.
Liu, H., Zhu, F., Yong, J., et al. (2008). Generation of induced pluripotent stem cells from adult rhesus monkey fibroblasts. Cell Stem Cell, 3, 587.
Ezashi, T., Telugu, B. P., Alexenko, A. P., Sachdev, S., Sinha, S., & Roberts, R. M. (2009). Derivation of induced pluripotent stem cells from pig somatic cells. Proceedings of the National Academy of Sciences of the United States of America, 106, 10993.
Kuo, C. H., Deng, J. H., Deng, Q., & Ying, S. Y. (2012). A novel role of miR-302/367 in reprogramming. Biochemical and Biophysical Research Communications, 417, 11.
Anokye-Danso, F., Trivedi, C. M., Juhr, D., et al. (2011). Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell, 8, 376.
Hussein, S. M., & Nagy, A. A. (2012). Progress made in the reprogramming field: new factors, new strategies and a new outlook. Current Opinion in Genetics & Development, 22, 435.
Vierbuchen, T., & Wernig, M. (2011). Direct lineage conversions: unnatural but useful? Nature Biotechnology, 29, 892.
Weintraub, H., Davis, R., Tapscott, S., et al. (1991). The myoD gene family: nodal point during specification of the muscle cell lineage. Science, 251, 761.
Najm, F. J., Lager, A. M., Zaremba, A., et al. (2013). Transcription factor-mediated reprogramming of fibroblasts to expandable, myelinogenic oligodendrocyte progenitor cells. Nature Biotechnology, 31, 426.
Yang, N., Zuchero, J. B., Ahlenius, H., et al. (2013). Generation of oligodendroglial cells by direct lineage conversion. Nature Biotechnology, 31, 434.
Vierbuchen, T., Ostermeier, A., Pang, Z. P., Kokubu, Y., Sudhof, T. C., & Wernig, M. (2010). Direct conversion of fibroblasts to functional neurons by defined factors. Nature, 463, 1035.
Szabo, E., Rampalli, S., Risueno, R. M., et al. (2010). Direct conversion of human fibroblasts to multilineage blood progenitors. Nature, 468, 521.
Qian, L., Huang, Y., Spencer, C. I., et al. (2012). In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature, 485, 593.
Song, K., Nam, Y. J., Luo, X., et al. (2012). Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature, 485, 599.
Gonzalez, F., Boue, S., & Izpisua Belmonte, J. C. (2011). Methods for making induced pluripotent stem cells: reprogramming a la carte. Nature Reviews Genetics, 12, 231.
Nair, V. (2008). Retrovirus-induced oncogenesis and safety of retroviral vectors. Current Opinion in Molecular Therapeutics, 10, 431.
Carey, B. W., Markoulaki, S., Hanna, J., et al. (2009). Reprogramming of murine and human somatic cells using a single polycistronic vector. Proceedings of the National Academy of Sciences of the United States of America, 106, 157.
Somers, A., Jean, J. C., Sommer, C. A., et al. (2010). Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem Cells, 28, 1728.
Zhou, W., & Freed, C. R. (2009). Adenoviral gene delivery can reprogram human fibroblasts to induced pluripotent stem cells. Stem Cells, 27, 2667.
Woltjen, K., Michael, I. P., Mohseni, P., et al. (2009). piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature, 458, 766.
Kaji, K., Norrby, K., Paca, A., Mileikovsky, M., Mohseni, P., & Woltjen, K. (2009). Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature, 458, 771.
Okita, K., Matsumura, Y., Sato, Y., et al. (2011). A more efficient method to generate integration-free human iPS cells. Nature Methods, 8, 409.
Yu, J., Chau, K. F., Vodyanik, M. A., Jiang, J., & Jiang, Y. (2011). Efficient feeder-free episomal reprogramming with small molecules. PLoS One, 6, e17557.
Si-Tayeb, K., Noto, F. K., Sepac, A., et al. (2010). Generation of human induced pluripotent stem cells by simple transient transfection of plasmid DNA encoding reprogramming factors. BMC Developmental Biology, 10, 81.
Zhou, H., Wu, S., Joo, J. Y., et al. (2009). Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell, 4, 381.
Kim, D., Kim, C. H., Moon, J. I., et al. (2009). Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell, 4, 472.
Fusaki, N., Ban, H., Nishiyama, A., Saeki, K., & Hasegawa, M. (2009). Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proceedings of the Japan Academy. Series B, Physical and Biological Sciences, 85, 348.
Warren, L., Manos, P. D., Ahfeldt, T., et al. (2010). Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell, 7, 618.
Yu, J., Vodyanik, M. A., Smuga-Otto, K., et al. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science, 318, 1917.
Luningschror P, Hauser S, Kaltschmidt B, Kaltschmidt C (2013) MicroRNAs in pluripotency, reprogramming and cell fate induction. Biochimica et Biophysica Acta
Subramanyam, D., Lamouille, S., Judson, R. L., et al. (2011). Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nature Biotechnology, 29, 443.
Liao, B., Bao, X., Liu, L., et al. (2011). MicroRNA cluster 302–367 enhances somatic cell reprogramming by accelerating a mesenchymal-to-epithelial transition. Journal of Biological Chemistry, 286, 17359.
Miyoshi, N., Ishii, H., Nagano, H., et al. (2011). Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell, 8, 633.
Yang, C. S., Li, Z., & Rana, T. M. (2011). microRNAs modulate iPS cell generation. RNA, 17, 1451.
Choi, Y. J., Lin, C. P., Ho, J. J., et al. (2011). miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nature Cell Biology, 13, 1353.
Wang, J., He, Q., Han, C., et al. (2012). p53-facilitated miR-199a-3p regulates somatic cell reprogramming. Stem Cells, 30, 1405.
Yang, C. S., & Rana, T. M. (2013). Learning the molecular mechanisms of the reprogramming factors: let's start from microRNAs. Molecular BioSystems, 9, 10.
Takahashi, K., Tanabe, K., Ohnuki, M., et al. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell, 131, 861.
Lowry, W. E., Richter, L., Yachechko, R., et al. (2008). Generation of human induced pluripotent stem cells from dermal fibroblasts. Proceedings of the National Academy of Sciences of the United States of America, 105, 2883.
Bao, L., He, L., Chen, J., et al. (2011). Reprogramming of ovine adult fibroblasts to pluripotency via drug-inducible expression of defined factors. Cell Research, 21, 600.
Maherali, N., Ahfeldt, T., Rigamonti, A., Utikal, J., Cowan, C., & Hochedlinger, K. (2008). A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell, 3, 340.
Stadtfeld, M., Maherali, N., Breault, D. T., & Hochedlinger, K. (2008). Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell, 2, 230.
Sauer, B., & Henderson, N. (1988). Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proceedings of the National Academy of Sciences of the United States of America, 85, 5166.
Chang, C. W., Lai, Y. S., Pawlik, K. M., et al. (2009). Polycistronic lentiviral vector for "hit and run" reprogramming of adult skin fibroblasts to induced pluripotent stem cells. Stem Cells, 27, 1042.
Sauer, B. (1992). Identification of cryptic lox sites in the yeast genome by selection for Cre-mediated chromosome translocations that confer multiple drug resistance. Journal of Molecular Biology, 223, 911.
Cochrane, R. L., Clark, S. H., Harris, A., & Kream, B. E. (2007). Rearrangement of a conditional allele regardless of inheritance of a Cre recombinase transgene. Genesis, 45, 17.
Yusa, K., Rad, R., Takeda, J., & Bradley, A. (2009). Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nature Methods, 6, 363.
Fraser, M. J., Ciszczon, T., Elick, T., & Bauser, C. (1996). Precise excision of TTAA-specific lepidopteran transposons piggyBac (IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera. Insect Molecular Biology, 5, 141.
Grabundzija, I., Wang, J., Sebe, A., et al. (2013). Sleeping Beauty transposon-based system for cellular reprogramming and targeted gene insertion in induced pluripotent stem cells. Nucleic Acids Research, 41, 1829.
Kues, W. A., Herrmann, D., Barg-Kues, B., et al. (2013). Derivation and characterization of sleeping beauty transposon-mediated porcine induced pluripotent stem cells. Stem Cells and Development, 22, 124.
Newman, J. C., Bailey, A. D., Fan, H. Y., Pavelitz, T., & Weiner, A. M. (2008). An abundant evolutionarily conserved CSB-PiggyBac fusion protein expressed in Cockayne syndrome. PLoS Genetics, 4, e1000031.
Ivics, Z., Hackett, P. B., Plasterk, R. H., & Izsvak, Z. (1997). Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell, 91, 501.
Stadtfeld, M., Nagaya, M., Utikal, J., Weir, G., & Hochedlinger, K. (2008). Induced pluripotent stem cells generated without viral integration. Science, 322, 945.
Okita, K., Nakagawa, M., Hyenjong, H., Ichisaka, T., & Yamanaka, S. (2008). Generation of mouse induced pluripotent stem cells without viral vectors. Science, 322, 949.
Yu, J., Hu, K., Smuga-Otto, K., et al. (2009). Human induced pluripotent stem cells free of vector and transgene sequences. Science, 324, 797.
Okita, K., Yamakawa, T., Matsumura, Y., et al. (2013). An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells, 31, 458.
Su, R. J., Baylink, D. J., Neises, A., et al. (2013). Efficient Generation of Integration-Free iPS Cells from Human Adult Peripheral Blood Using BCL-XL Together with Yamanaka Factors. PLoS One, 8, e64496.
Jia, F., Wilson, K. D., Sun, N., et al. (2010). A nonviral minicircle vector for deriving human iPS cells. Nature Methods, 7, 197.
Narsinh, K. H., Jia, F., Robbins, R. C., Kay, M. A., Longaker, M. T., & Wu, J. C. (2011). Generation of adult human induced pluripotent stem cells using nonviral minicircle DNA vectors. Nature Protocols, 6, 78.
Li, H. O., Zhu, Y. F., Asakawa, M., et al. (2000). A cytoplasmic RNA vector derived from nontransmissible Sendai virus with efficient gene transfer and expression. Journal of Virology, 74, 6564.
Nagai, Y., & Kato, A. (2004). Accessory genes of the paramyxoviridae, a large family of nonsegmented negative-strand RNA viruses, as a focus of active investigation by reverse genetics. Current Topics in Microbiology and Immunology, 283, 197.
Shioda, T., Iwasaki, K., & Shibuta, H. (1986). Determination of the complete nucleotide sequence of the Sendai virus genome RNA and the predicted amino acid sequences of the F, HN and L proteins. Nucleic Acids Research, 14, 1545.
Takimoto, T., Taylor, G. L., Connaris, H. C., Crennell, S. J., & Portner, A. (2002). Role of the hemagglutinin-neuraminidase protein in the mechanism of paramyxovirus-cell membrane fusion. Journal of Virology, 76, 13028.
Masaki, I., Yonemitsu, Y., Komori, K., et al. (2001). Recombinant Sendai virus-mediated gene transfer to vasculature: a new class of efficient gene transfer vector to the vascular system. FASEB journal : official publication of the Federation of American Societies for Experimental Biology, 15, 1294.
Yonemitsu, Y., Kitson, C., Ferrari, S., et al. (2000). Efficient gene transfer to airway epithelium using recombinant Sendai virus. Nature Biotechnology, 18, 970.
Murakami, Y., Ikeda, Y., Yonemitsu, Y., et al. (2008). Newly-developed Sendai virus vector for retinal gene transfer: reduction of innate immune response via deletion of all envelope-related genes. The Journal of Gene Medicine, 10, 165.
Nishimura, K., Sano, M., Ohtaka, M., et al. (2011). Development of defective and persistent Sendai virus vector: a unique gene delivery/expression system ideal for cell reprogramming. Journal of Biological Chemistry, 286, 4760.
Hsu, M., Scheid, A., & Choppin, P. W. (1981). Activation of the Sendai virus fusion protein (f) involves a conformational change with exposure of a new hydrophobic region. Journal of Biological Chemistry, 256, 3557.
Mizumoto, K., Muroya, K., Takagi, T., Omata-Yamada, T., Shibuta, H., & Iwasaki, K. (1995). Protein factors required for in vitro transcription of Sendai virus genome. Journal of Biochemistry, 117, 527.
Garcin, D., Pelet, T., Calain, P., Roux, L., Curran, J., & Kolakofsky, D. (1995). A highly recombinogenic system for the recovery of infectious Sendai paramyxovirus from cDNA: generation of a novel copy-back nondefective interfering virus. The EMBO Journal, 14, 6087.
Hasan, M. K., Kato, A., Shioda, T., Sakai, Y., Yu, D., & Nagai, Y. (1997). Creation of an infectious recombinant Sendai virus expressing the firefly luciferase gene from the 3' proximal first locus. The Journal of General Virology, 78(Pt 11), 2813.
Inoue, M., Tokusumi, Y., Ban, H., et al. (2003). Nontransmissible virus-like particle formation by F-deficient sendai virus is temperature sensitive and reduced by mutations in M and HN proteins. Journal of Virology, 77, 3238.
Nishishita, N., Takenaka, C., Fusaki, N., & Kawamata, S. (2011). Generation of human induced pluripotent stem cells from cord blood cells. Journal of Stem Cells, 6, 101.
Seki, T., Yuasa, S., Oda, M., et al. (2010). Generation of induced pluripotent stem cells from human terminally differentiated circulating T cells. Cell Stem Cell, 7, 11.
Seki, T., Yuasa, S., & Fukuda, K. (2012). Generation of induced pluripotent stem cells from a small amount of human peripheral blood using a combination of activated T cells and Sendai virus. Nature Protocols, 7, 718.
Ban, H., Nishishita, N., Fusaki, N., et al. (2011). Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. Proceedings of the National Academy of Sciences of the United States of America, 108, 14234.
Nakanishi, M., & Otsu, M. (2012). Development of Sendai virus vectors and their potential applications in gene therapy and regenerative medicine. Current Gene Therapy, 12, 410.
Griesenbach, U., Inoue, M., Meng, C., et al. (2012). Assessment of F/HN-pseudotyped lentivirus as a clinically relevant vector for lung gene therapy. American Journal of Respiratory and Critical Care Medicine, 186, 846.
Masaki, I., Yonemitsu, Y., Yamashita, A., et al. (2002). Angiogenic gene therapy for experimental critical limb ischemia: acceleration of limb loss by overexpression of vascular endothelial growth factor 165 but not of fibroblast growth factor-2. Circulation Research, 90, 966.
Kano, M., Matano, T., Nakamura, H., et al. (2000). Elicitation of protective immunity against simian immunodeficiency virus infection by a recombinant Sendai virus expressing the Gag protein. AIDS, 14, 1281.
Griesenbach, U., Inoue, M., Hasegawa, M., & Alton, E. W. (2005). Sendai virus for gene therapy and vaccination. Current Opinion in Molecular Therapeutics, 7, 346.
Grudzien-Nogalska, E., Kowalska, J., Su, W., et al. (2013). Synthetic mRNAs with superior translation and stability properties. Methods in Molecular Biology, 969, 55.
Kariko, K., Buckstein, M., Ni, H., & Weissman, D. (2005). Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity, 23, 165.
Hornung, V., Ellegast, J., Kim, S., et al. (2006). 5'-Triphosphate RNA is the ligand for RIG-I. Science, 314, 994.
Roitsch, T., & Lehle, L. (1989). Requirements for efficient in vitro transcription and translation: a study using yeast invertase as a probe. Biochimica et Biophysica Acta, 1009, 19.
Anderson, B. R., Muramatsu, H., Nallagatla, S. R., et al. (2010). Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Research, 38, 5884.
Darzynkiewicz, E., Stepinski, J., Ekiel, I., et al. (1988). Beta-globin mRNAs capped with m7G, m2.7(2)G or m2.2.7(3)G differ in intrinsic translation efficiency. Nucleic Acids Research, 16, 8953.
Jemielity, J., Fowler, T., Zuberek, J., et al. (2003). Novel "anti-reverse" cap analogs with superior translational properties. RNA, 9, 1108.
Pichlmair, A., Schulz, O., Tan, C. P., et al. (2006). RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science, 314, 997.
Kariko, K., & Weissman, D. (2007). Naturally occurring nucleoside modifications suppress the immunostimulatory activity of RNA: implication for therapeutic RNA development. Current Opinion in Drug Discovery & Development, 10, 523.
Kariko, K., Muramatsu, H., Welsh, F. A., et al. (2008). Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Molecular Therapy : The Journal of the American Society of Gene Therapy, 16, 1833.
Motorin, Y., & Helm, M. (2011). RNA nucleotide methylation. Wiley Interdisciplinary Reviews RNA, 2, 611.
Mandal, P. K., & Rossi, D. J. (2013). Reprogramming human fibroblasts to pluripotency using modified mRNA. Nature Protocols, 8, 568.
Kormann, M. S., Hasenpusch, G., Aneja, M. K., et al. (2011). Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nature Biotechnology, 29, 154.
Kariko, K., Muramatsu, H., Keller, J. M., & Weissman, D. (2012). Increased erythropoiesis in mice injected with submicrogram quantities of pseudouridine-containing mRNA encoding erythropoietin. Molecular Therapy : The Journal of The American Society of Gene Therapy, 20, 948.
Harris K, Aylott M, Cui Y, Louttit JB, McMahon NC, Sridhar A (2013) Comparison of electrophysiological data from human induced pluripotent stem cell derived cardiomyocytes (hiPSC-CMs) to functional pre-clinical safety assays. Toxicological sciences : an official journal of the Society of Toxicology
Gai, H., Leung, E. L., Costantino, P. D., et al. (2009). Generation and characterization of functional cardiomyocytes using induced pluripotent stem cells derived from human fibroblasts. Cell Biology International, 33, 1184.
Zhang, J., Wilson, G. F., Soerens, A. G., et al. (2009). Functional cardiomyocytes derived from human induced pluripotent stem cells. Circulation Research, 104, e30.
Ma, J., Guo, L., Fiene, S. J., et al. (2011). High purity human-induced pluripotent stem cell-derived cardiomyocytes: electrophysiological properties of action potentials and ionic currents. American Journal of Physiology - Heart and Circulatory Physiology, 301, H2006.
Addis RC, Ifkovits JL, Pinto F, et al. (2013) Optimization of direct fibroblast reprogramming to cardiomyocytes using calcium activity as a functional measure of success. Journal of Molecular and Cellular Cardiology
Efe, J. A., Hilcove, S., Kim, J., et al. (2011). Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nature Cell Biology, 13, 215.
Graf, T., & Enver, T. (2009). Forcing cells to change lineages. Nature, 462, 587.
Zhou, Q., Brown, J., Kanarek, A., Rajagopal, J., & Melton, D. A. (2008). In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature, 455, 627.
Sekiya, S., & Suzuki, A. (2011). Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature, 475, 390.
Papp, B., & Plath, K. (2011). Reprogramming to pluripotency: stepwise resetting of the epigenetic landscape. Cell Research, 21, 486.
Zwi, L., Caspi, O., Arbel, G., et al. (2009). Cardiomyocyte differentiation of human induced pluripotent stem cells. Circulation, 120, 1513.
Ieda, M., Fu, J. D., Delgado-Olguin, P., et al. (2010). Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell, 142, 375.
Papapetrou, E. P., Tomishima, M. J., Chambers, S. M., et al. (2009). Stoichiometric and temporal requirements of Oct4, Sox2, Klf4, and c-Myc expression for efficient human iPSC induction and differentiation. Proceedings of the National Academy of Sciences of the United States of America, 106, 12759.
Hirai, H., Tani, T., Katoku-Kikyo, N., et al. (2011). Radical acceleration of nuclear reprogramming by chromatin remodeling with the transactivation domain of MyoD. Stem Cells, 29, 1349.
Hirai, H., Katoku-Kikyo, N., Karian, P., Firpo, M., & Kikyo, N. (2012). Efficient iPS cell production with the MyoD transactivation domain in serum-free culture. PLoS One, 7, e34149.
Acknowledgments
I thank Marta Roche, Dr. Grahame McKenzie, Dr. Giovanna Giovinazzo, and the members of the Stem Cell Biology Program for the helpful discussion. This work was supported by a grant (BFU2012-35258) to J.A. Bernal from the Ministry of Economy and Competitivity. J.A. Bernal was also supported by the Spanish Ramón y Cajal program (RYC2009-04341) from the Ministry of Science and Innovation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Associate Editor Enrique Lara-Pezzi oversaw the review of this article
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Bernal, J.A. RNA-Based Tools for Nuclear Reprogramming and Lineage-Conversion: Towards Clinical Applications. J. of Cardiovasc. Trans. Res. 6, 956–968 (2013). https://doi.org/10.1007/s12265-013-9494-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12265-013-9494-8