Abstract
Uncovering the genetic risk and protective factors for complex diseases is of fundamental importance for advancing therapeutic and biomarker discoveries. This endeavor is particularly challenging for neuropsychiatric diseases where diagnoses predominantly rely on the clinical presentation, which may be heterogeneous, possibly due to the heterogeneity of the underlying genetic susceptibility factors and environmental exposures. Although genome-wide association studies of various neuropsychiatric diseases have recently identified susceptibility loci, there likely remain additional genetic risk factors that underlie the liability to these conditions. Furthermore, identification and characterization of the causal risk variant(s) in each of these novel susceptibility loci constitute a formidable task, particularly in the absence of any prior knowledge about their function or mechanism of action. Biologically relevant, quantitative phenotypes, i.e., endophenotypes, provide a powerful alternative to the more traditional, binary disease phenotypes in the discovery and characterization of susceptibility genes for neuropsychiatric conditions. In this review, we focus on Alzheimer’s disease (AD) as a model neuropsychiatric disease and provide a synopsis of the recent literature on the use of endophenotypes in AD genetics. We highlight gene expression, neuropathology and cognitive endophenotypes in AD, with examples demonstrating the utility of these alternative approaches in the discovery of novel susceptibility genes and pathways. In addition, we discuss how these avenues generate testable hypothesis about the pathophysiology of genetic factors that have far-reaching implications for therapies.
Similar content being viewed by others
Introduction
Genetic studies of human diseases have been marked by an explosion in the number of susceptibility loci identified through genome wide association studies (GWAS) in the past several years. Similar to other complex disorders, neuropsychiatric diseases, too, had a share of their genetic risk loci discoveries, with—for example—28 published studies to date on Alzheimer’s disease (AD), 18 on bipolar disorder and 22 on schizophrenia, according to the Catalog of Published GWAS accessed on 21 Aug 2012 [1]. The translation of this success to viable therapies and biomarker discovery depends on the identification and characterization of the actual disease genes and functional variants at these susceptibility loci. Furthermore, despite the large number of discovered loci, a substantial component of genetic susceptibility remains unexplained for complex diseases [2•]. To overcome these major hurdles in the post-GWAS era requires a multitude of alternative approaches including the use of endophenotypes, which are biologically-relevant, quantitative and heritable phenotypes [3•] (Fig. 1a).
The endophenotype approach was initially advocated in psychiatric genetics [4, 5] due to the need to have an objective and quantifiable outcome in genetic studies, given the relatively imprecise nature of the clinical diagnosis, which is thought to result in heterogeneity. The rationale for using endophenotypes, instead of or in addition to the binary disease phenotypes, stems from the following assumptions: (1) Endophenotypes represent an intermediate outcome between genes and clinical diagnosis of a disease, and given their closer proximity to the genetic variation than the disease outcome, the genetic component influencing the endophenotype will be larger and therefore easier to detect [6, 7]; (2) Endophenotypes are under the influence of a smaller number of genes than the more complex disease outcome [6]; (3) Given its quantitative nature, using an endophenotype as an outcome variable will be statistically more powerful than the binary case/control approach in detecting genetic associations; (4) Since they are objectively quantifiable, endophenotypes constitute a more homogeneous and accurately measurable phenotype than disease outcome; (5) The endophenotype approach can allow inclusion of individuals with and without a given diagnosis, which will increase power, particularly in family studies or for traits that are age dependent [7, 8]; (6) The endophenotype approach will provide information about the underlying mechanism of action for the gene and variant of interest and might therefore more readily enable downstream functional investigations, including the generation of animal models with quantifiable outcomes [9]. While the accuracy of these assumptions needs to be established, the endophenotype approach has begun to generate hypotheses for novel genetic loci and pathways implicated in human disease and to enable the downstream characterization of disease variants and genes, as exemplified in this review (Fig. 1b).
AD, the most common dementia in the elderly, is especially amenable to the endophenotype approach, for a number of reasons. First, AD has a distinct neuropathology characterized by accumulation of amyloid β (Aβ) in senile plaques and hyperphosphorylated tau in neurofibrillary tangles, both of which are quantifiable phenotypes, and the latter correlates with clinical disease severity [10]. Second, discoveries of Mendelian mutations in the amyloid precursor protein (APP), presenilin 1 (PSEN1) and PSEN2 genes in early-onset AD that lead to elevations in secreted Aβ and their modeling in animals harboring these mutations (reviewed [11]) bolstered the amyloid cascade hypothesis [12]. The ability to measure Aβ levels in the serum and cerebrospinal fluid (CSF) of AD patients and their relatives [13], and the determination that Aβ levels are heritable [14] enabled the first studies utilizing Aβ levels as endophenotypes in genetic studies that discovered genetic loci and variants influencing AD risk and Aβ [8, 15–17]. This was followed by investigations of CSF Aβ [18] and tau levels [19] as endophenotypes in AD genetics studies. Third, the availability of prospective, elderly cohorts with rich clinical, neurocognitive and neuroimaging measures [20–22], knowledge that many of these measures are heritable [6, 7, 23], and detection of preclinical changes in these measures (reviewed [24••]) advocate their use as endophenotypes in genetic studies of AD. Fourth, the advent of technology that allows measurement of gene expression levels for all known transcripts (transcriptome), development of methodologies that allow analysis of this data at the whole-genome level, significant heritability attributed to gene expression levels (reviewed [3•]) and the availability of well-characterized brain tissue from neuropathologic AD and other patients in which transcriptome can be measured, empowered the use of gene expression levels from brain and other tissues as endophenotypes in AD.
This review focuses on three types of endophenotypes in AD: gene expression levels, neuropathologic measures and cognitive measures. These diverse endophenotypes span the vast spectrum of biological insights that can be gained by this quantitative approach: genetic associations with transcript levels, the most proximal of these traits to the susceptibility allele, may uncover the initial mechanism for the functional consequences of the allele. Neuropathologic phenotypes can relate a variant to the known neuropathology of the disease and might enable the dissection of pathophysiologic pathways influenced by the polymorphism of interest. Finally, the use of cognitive endophenotypes can uncover genetic risk factors governing distinct aspects of human cognition and the clinical expression of the disease. We recognize that there are many other endophenotypes that are currently utilized or are excellent candidates for genetic studies of AD, including Aβ and tau levels, neuroimaging measures such as hippocampal volume and magnetic resonance spectroscopy levels and methylation patterns. Although a comprehensive assessment of all these endophenotypes is beyond the scope of this review, the generalizations that can be drawn from this synopsis could potentially be applicable to many other quantitative phenotypes in AD research.
Gene Expression Endophenotype
Gene expression levels constitute a special group of endophenotypes for a number of reasons. First, because the tested phenotype is the level of the expressed gene transcript(s), genetic studies of gene expression endophenotypes (also known as expression quantitative trait loci or eQTL studies) directly identify the gene under the influence of genetic variants. This is in contrast to any other phenotype, where genetic studies merely implicate a “locus of interest” without definitive information about the affected gene. This first characteristic of the gene expression endophenotype can be utilized to uncover plausible disease genes via combined assessment of gene expression endophenotype and disease phenotype as discussed below. Second, genetic factors identified via eQTL studies provide guidance about the underlying mechanism of action of the “functional variants” at the “locus of interest”. This can enable a more directed search for such “functional variants”, for example by focusing on regulatory variants that influence whole transcript levels or splice isoforms. Furthermore, such information can guide downstream in vitro studies that are more relevant to the underlying genetic variation. Third, the ability to assess concerted expression level changes or eQTL associations at the transcriptome level via pathway analysis can lead to identification of novel biological networks that may underlie disease pathophysiology. Below, we discuss the utility of gene expression endophenotypes in gene discovery and characterization in AD, highlighting examples that take advantage of these special characteristics of this approach.
Utility in Gene Discovery
The utilization of gene expression endophenotypes in gene discovery in AD first began with transcriptome profiling (or mRNA profiling) studies, which are recently comprehensively reviewed [25]. The underlying premise of these studies is that mRNA from patients with disease will have changes in comparison to controls; and that these changes may underlie disease pathophysiology. The most important caveat in this assumption is that the detected gene expression changes may be a consequence of the disease and non-specific, rather than a causal event [3•]. This pitfall is especially concerning if the transcriptome profiling is performed in tissue affected by the disease (such as the temporal cortex in AD). Indeed, in a detailed microarray-based transcriptome profiling study of 14 different cerebral cortex regions and the hippocampus, from 69 autopsied AD subjects of varying clinical and pathologic severity versus 18 controls (maximal number of subjects utilized in the study), Haroutunian et al. [26] identified the greatest number of gene expression changes in regions from the temporal cortex across the disease stages, with increasing changes occurring in later disease stages and stronger correlations between gene expression and disease severity seen in more advanced disease. Importantly, most of the changes observed were downregulations rather than upregulations. Collectively, these results could imply that the progression of disease and cell loss may be driving these changes, rather than vice versa. These authors [26] and others [27] attempted to overcome this concern by analysis of autopsied AD subjects with mild neuropathology and concluded that gene expression changes that occur in regions prior to the development of neuropathology are unlikely to be a consequence of the disease process. Bossers et al. [27] analyzed 49 prefrontal cortex samples from subjects with Alzheimer’s-type neuropathology to identify correlated changes in gene expression which varied with advancing Braak stage. They determined that the most significant changes occurred in “synaptic activity genes” between Braak stages II and III, which is prior to or just at the onset of AD-type neuropathology when the subjects were clinically non-demented. The authors also noted that levels of several genes correlated with increasing intracellular Aβ levels during these Braak stages, leading them to postulate that expression changes in genes of synaptic activity may be a coping mechanism against increased Aβ that occurs prior to clinical and neuropathological AD. While it is not possible to draw definitive conclusions about the longitudinal cascade of events, including gene expression changes, based on cross-sectional assessment of brain tissue from small numbers of distinct subjects, these results nevertheless generate intriguing hypotheses about AD pathophysiology via correlative analysis of transcriptome and neuropathology data.
Another approach in utilizing gene expression endophenotypes in gene discovery is combined transcriptome profiling and AD risk association studies. In a small hippocampal mRNA profiling study of six AD versus two control brains, Li et al. [15] detected lower GSTO1 (glutathione S-transferase omega-1) levels in the AD brains, followed by significant associations with age-at-onset of both AD and Parkinson’s disease with variants in both GSTO1 and its nearby homologue GSTO2 [28]. This prompted follow-up genetic studies with disease risk and/or age-at-onset phenotypes with mixed results [29–33]. In a GWAS of brain gene expression (brain eGWAS) levels in ~800 tissue samples from ~400 brains [34•], we identified strong associations with variants at this locus and brain GSTO2 but not GSTO1 levels [35], consistent with results from another brain eQTL study [36]. In our study, we determined that the same variant associated with both lower brain levels of GSTO2 as well as increased AD risk in older subjects, which is biologically consistent with the antioxidant functions of this gene. Furthermore, pathway analysis of the significant genes in our brain eGWAS showed significant enrichment for glutathione metabolism genes, suggesting there may be additional genes in this pathway with potential influence on AD and other neurodegenerative diseases. Other genes which were detected by expression profiling studies of AD versus control tissue, followed by significant associations with AD risk, include POU2F1 [37] and IL-33 [38]. These studies highlight the potential utility of the gene expression endophenotype in identifying gene(s) and pathways that may harbor regulatory variants that influence disease risk.
More recently, joint assessment of disease GWAS with eQTL studies have been advocated to prioritize suggestive results from disease GWAS and/or identify novel candidate disease genes, based on the premise that disease variants will be enriched for regulatory variants that influence gene expression and vice versa [39••]. Indeed, in a comparison of eQTL results from lymphoblastoid cell lines from HapMap samples with human disease/trait GWAS summary data, Nicolae et al. [40] identified significant enrichment for SNPs that influence expression (eSNPs) amongst human disease/trait associating variants. Combined assessment of brain expression endophenotype associations [36] with disease GWAS showed enrichment for eQTLs amongst schizophrenia risk alleles [41]. This approach, combined with pathway analysis led to nomination of novel genes for diabetes in another study [42]. We have applied this approach for the first time to a large AD GWAS [43], by combining with our brain eGWAS data and detected an enrichment for significant eSNPs amongst suggestive AD risk SNPs [34•]. These results suggest that AD, like other complex human diseases, may at least in part be influenced by regulatory variants. The novel genes detected in the “grey zone” of disease GWAS by this approach warrant further studies for identification of functional variants and to demonstrate their downstream regulatory effects.
Utility in Gene Characterization
Gene expression endophenotypes can also be used to characterize the effects of disease risk variants and their downstream consequences on the disease gene. A prime example of this is MAPT, which has rare variants leading to frontotemporal dementia with parkinsonism linked to chromosome 17, as well as common variants within a haplotype block that associate with multiple taupathies (reviewed [44]). These variants have been shown to influence either splicing or transcriptional activity of MAPT [45–48].
Another example of gene expression endophenotype explorations for a known risk gene is APOE, which has common missense polymorphisms, leading to three isoforms APOE ε2, ε3 or ε4, where APOE ε4 has clearly been shown to influence AD risk, whereas APOE ε2 might confer protection from AD [11, 49]. APOE isoforms have dual types of functions in the brain with roles in both maintaining neural health and also in promoting AD pathophysiology (reviewed [50]). In addition to the most well-studied isoforms, a number of promoter region polymorphisms have been identified for APOE that impart risk for AD, at least partially independently of the APOE isoform (reviewed [51]). Although consensus is still lacking, the most well studied promoter region polymorphism −491AA appears to confer AD risk independent of APOE ε4 and increase APOE transcriptional activity [52, 53], suggesting that both APOE isoforms and levels may play a role in AD pathogenicity. Despite absence of conclusive evidence for the role of the promoter region polymorphisms in AD risk, the transcriptional complexity of APOE and its dual role in the central nervous system (CNS), APOE-directed therapeutics aimed at modifying its levels are advocated for treatment of AD. A recent study in animal models of AD demonstrated clearing of Aβ and reversal of behavioral and electrophysiologic deficits upon treatment with a transcriptional inducer of APOE [54].
It is similarly critical to characterize the novel AD candidate variants and genes that are being identified in late-onset AD (LOAD) GWAS (reviewed [55]) with respect to their influence on gene expression endophenotypes. This information will provide focus for the downstream functional variant discovery, in vitro and in vivo studies and ultimately set the stage for the search of therapeutics targeting the appropriate mechanisms and pathways. We have begun to characterize the novel LOAD GWAS variants for their influence on gene expression endophenotypes using our brain eGWAS [56•] and eQTL analyses of data generated from peripheral immune cells [57]. The latter study demonstrated that AD-associated variants, such as the one in the PICALM locus, influence gene expression in non-resident CNS cells and suggest that infiltrating immune cells may play a role in the onset of AD. On the other hand, the brain data identified association between the top AD risk variants at the CLU and MS4A loci with brain levels of CLU and MS4A4A genes, implicating regulatory genetic variation for these genes in AD risk. Furthermore, we detected additional strong gene expression associations for both CLU and ABCA7, some of which also confer AD risk, independent of the top GWAS variants, suggesting that new regulatory AD variants might exist at these loci, in addition to the top SNPs already identified by disease GWAS. Our findings in CLU are corroborated by Ling et al. [58•] who determined that the AD-protective CLU variant is also associated with higher CLU1 isoform levels in human brains. The direction of the gene expression endophenotype effect is identical in these two studies, and indicate that therapeutic approaches aimed at increasing levels of CLU in the brain might confer protection from AD. Interestingly, valproic acid (VPA), a well-known anti-epileptic and anti-depressant with histone deacetylase (HDAC) inhibiting properties, was shown to induce CLU expression in astrocytes [59]. VPA was also previously highlighted as a potentially promising drug for AD due to its pro-neurogenesis and neuroprotective properties [60]. Although clinical trials of VPA in AD patients have yet failed to demonstrate a beneficial outcome [61, 62], evidence warrants further investigations along the CLU induction axis as a potential therapeutic avenue in AD. Gene expression endophenotypes may be informative biomarkers in such future therapeutic trials.
Neuropathology Endophenotype
Despite recent achievements by various GWAS consortia [43, 63–66], a large proportion of the genetic contribution to Alzheimer disease still remains to be identified. The utility of the conventional approach that relies heavily on clinical diagnosis (i.e., a dichotomy of cases vs. controls) is dampened by contamination of the control group with persons with pre- or sub-clinical disease. Use of the intermediate neuropathologic endophenotype helps to address these disadvantages. While the majority of persons clinically diagnosed with AD have AD pathology [67–69], AD pathology is also common among persons without dementia [70–73]. Neuropathologic abnormality have been reported in persons both with and without cognitive impairment [74, 75], suggestive of a disease process involving pathologic change of brain structure. Moreover, the phenotypic heterogeneity of dementia reflects a broader spectrum of neurodegenerative conditions other than AD, including cerebrovascular infarctions, neocortical Lewy bodies, and TAR DNA-binding protein 43 (TDP-43), just to name a few. Each of these diseases independently contributes to the clinical dementia phenotype [76, 77]. Further, this heterogeneity extends to the probable AD phenotype [78]. Given this context, neuropathologic phenotypes provide several important implications.
First, compared to the more distal clinical phenotypes, neuropathologic traits lie directly in the pathway connecting genetic actions to the clinical expression of AD dementia. In other words, genetic variants do not directly cause cognitive decline and AD, but rather contribute to a series of events associated with neuropathology; these, in turn, result in cognitive decline and AD. Thus, utilization of neuropathologic outcomes increases statistical power to discover genetic variants that influence AD-related processes. Using the well-known apolipoprotein E genotype (APOE), we demonstrated that among a group of only about 500 community based elderly with European ancestry, quantitative pathologic AD phenotypes provide considerably more power than phenotypes of clinical AD diagnosis or cognitive function [79•]. In this analysis, the association of the protective APOE ε2 allele with clinical AD and level of cognition were not significant. However, it has a strong association with a measure of overall burden of AD pathology (p = 10 × 10−5). Similar differences were seen with the ε4 allele. We subsequently showed that measures of AD pathology mediated the association of allele status with cognitive decline illustrating that AD pathology is in the causal chain linking the genetic variant with cognitive decline [80]. Other studies have reported similar findings [81].
Second, genetic associations with clinical outcomes are confounded by misclassification of pre- and sub-clinical subjects. These are people that are harboring genetic variants that link to AD pathophysiology, but these persons have not yet reached the threshold for a clinical AD diagnosis. As a result, the magnitude of the association can be diluted due to the discordance between AD neuropathology and diagnostic status. For example, in the study outlined above, we found that APOE ε4 was associated with AD pathology among persons without dementia, i.e., in analyses restricted to the control group of a case–control study [79•]. This issue is further complicated by individual differences in cognitive or neural reserve [82]. Both structural [83] and neuropsychological [84] components of reserve have been shown to influence the level of resilience in the face of accumulating disease pathology, such that a greater reserve capacity reduces the deleterious effect of AD pathology on clinical symptoms. Without directly assessing genetic influences on disease pathology, such influences could be easily masked by the modifying effect of reserve. In a recent GWAS of AD pathology, Kramer et al. [85•] discovered that polymorphisms in RELN were associated with higher burden of neurofibrillary tangles (NFT) among older persons without dementia, and they hypothesized a potential role of reelin in tau phosphorylation and that upregulation of reelin may be a compensatory remedy to tau-related stress.
Third, a compelling rationale for using an endophenotype is to refine and partition a generic phenotype into ones that are associated with very specialized pathways [6]. The fine-tuning helps to address the complexity of the disease biology and can amplify the association of contributory loci along the targeted pathway. Substantial evidence shows that AD tends to be co-existent with other brain lesions like cerebrovascular infarctions, Lewy bodies, and TDP-43, suggesting that not all AD-associated alleles will work through the pathologic accumulation of Aβ and phosphorylation of tau, the pathologic hallmarks that characterize AD. The disease also involves many other biological processes such as oxidative stress [86], chronic inflammation [87], alteration in lipid metabolism [88] and depletion of molecular chaperones [89]. It is essential to disentangle distinct genetic risk factors for the different intermediate traits in order to understand the underlying biological mechanisms that contribute to the onset of AD. Sleegers et al. [90] presented a conceptual model for the implications of recently discovered loci on AD susceptibility highlighting the influence of these novel loci on different aspects of AD pathophysiology. Clusterin (CLU) was hypothesized to share many properties of APOE in regulating Aβ formation and lipid transportation; complement receptor 1 (CR1) on the other hand likely contributed to chronic inflammation and C3b-mediated clearance; and phosphatidylinositol binding clathrin assembly protein (PICALM) was implicated in maintaining synaptic function and mediating endocytosis in APP recycling.
Pathologic phenotypes offer the promise of assessing different mechanistic hypotheses of action for each risk allele. For example, we showed that CR1, but not CLU and PICALM, was significantly associated with deposition of neuritic plaques, and this association further mediates, in part, the effect of the CR1 locus on cognitive decline [91•]. However, the CR1 locus also affects the accumulation of cerebral amyloid angiopathy [92] and may therefore also function through an effect on the cerebral vasculature. Beyond demonstrating the utility of leveraging intermediate phenotypes to build a causal chain of events leading from a risk factor to a clinical syndrome, CR1 also illustrates the strategy of using an intermediate phenotype to perform fine mapping of a susceptibility locus, which can help to locate the causal variant and to find additional variants that have an effect on AD and its pathology [93]. These types of studies are not unique to APOE and CR1, as evidence supporting an association of the CETP AD susceptibility allele with AD pathology has recently been reported [94]. Further, an interesting study from Brazil reported that an individual’s proportion of African ancestry was associated with a lower burden of neuritic plaque pathology, although no specific variants were reported [95].
Cognition Endophenotype
In parallel to the neuropathologic phenotypes, cognitive endophenotypes (i.e., level of cognitive function and rate of decline in cognition) serve as another promising alternative in gene discovery [96]. These quantitative measures share the strengths of the neuropathologic phenotypes as presented above. In particular, they are independent of diagnostic status and can be assessed in persons with and without the clinical manifestation of the disease, which helps to overcome the obstacle of confounding due to pre- and sub-clinical contamination in the control group. We and others have shown that cognitive decline begins years prior to a clinical diagnosis of AD or MCI [97]. Further, because of their quantitative nature, statistical power to capture heritable variation is improved. It is now clear that a better understanding of the earlier stage in the disease progression holds great promise for effective prevention and intervention strategies, and cognitive phenotypes can help to detect genetic risk factors attributable to the preclinical and subclinical change in cognition that are not likely to be captured in conventional case–control studies.
An important additional strength of cognitive endophenotypes that complements neuropathologic traits is that they can be measured within the individuals longitudinally throughout life. Compared with cross-sectional data, these longitudinal data directly address the question of change over time [98]. AD is the result of a sequence of pathophysiological events from Aβ deposition to synaptic dysfunction, to tangle formation, to other structural changes [99••]. Trajectories of change in cognition characterized by repeated assessments of cognition provide objective evidence about how AD manifests over time. Endophenotypes such as cognitive decline have been increasingly used [91•, 93, 94, 96, 100•, 101–107] to explore the genetic linkage to these trajectories in two important ways.
First, the AD loci discovered so far in cross-sectional susceptibility studies are interestingly not in concordance with those that account for the disease progression. In a recent genome-wide scan, none of the known AD susceptibility variants, except APOE and CR1, were found to be significantly associated with the rate of cognitive decline [100•]. On the other hand, the study reported a highly suggestive association with a locus near the PDE7A and MTFR1 genes which regulate inflammation and oxidative stress, respectively. Similar non-findings were reported by a separate GWAS effort, where minor allele homozygosity of multiple novel variants were found to be associated with a faster rate of disease progression in subjects with mild cognitive impairment, but none of these variants matched those identified in previous susceptibility studies [107]. However, it should be noted that the sample sizes of these cognitive decline GWAS are a fraction of the size of the case/control studies, so it is too early to make definitive statements about known and novel susceptibility loci. Estimates suggest that >5,000 subjects will be needed to begin to have reasonable power to identify a given variant [100•].
Second, AD develops slowly over decades and the cognitive trajectories cover a wide spectrum from preclinical phase of AD [108] all the way through the terminal decline in the last few years of life [109]. Specific AD susceptibility loci could be associated with different aspects of this cascade; therefore, it is plausible that they differentially affect various stages of the cognitive trajectory. Intermediate phenotypes like cognitive decline provide additional utility in dissecting the functional pathway in gene action. APOE is again illustrative on this point: the AD susceptibility allele APOE ε4 was discovered decades ago, and so far little is known regarding where the polymorphism exerts its effect over the course of the disease. In particular, it is not clear whether the effect of the APOE locus persists after the onset of dementia or whether it differs in its magnitude of influence along the progression. Most literature consistently reports the effect of ε4 on the risk of incident AD [110–113] and decline in cognitive performance in persons free of dementia [102, 114–117, 118•, 119, 120]. Controversy arises on whether there is an ε4 effect on cognitive decline in the late stages of the disease. Some studies suggest that ε4 is not related to decline after a diagnosis of AD [121–125], which would support the theory that APOE works primarily as a triggering factor [126]. On the other hand, other studies have found that the ε4 allele remains as an important predictor of the progression to AD after subjects experience cognitive impairment [127, 128] and is associated with cognitive decline in the early stages of AD [129]. To unravel these controversies, more complex analyses such as nonlinear mixed models could be considered for studies that have a sufficient number of cognitive evaluations over a long enough time [130]. Recently, using random change point models, we have showed that, among participants who were dementia free at enrollment but later developed incident AD, ε4 carriers had a more rapid cognitive decline both before and after the onset of AD dementia [80]. The capacity to incorporate these types of analyses into high-throughput gene discovery programs will best exploit the use of endophenotypes derived from longitudinal data for GWAS.
Future Directions
As the meta-analyses of AD GWAS come to a close, we will have a number of validated and suggested susceptibility loci whose functional consequences can begin to be elucidated by leveraging pertinent intermediate traits such as the ones that we have discussed: RNA expression, neuropathologic measures and cognitive measures (Fig. 2). As illustrated by the APOE and CR1 loci, such studies can be critical in tying risk factors to a particular aspect of AD-related pathophysiology and can lead to the elaboration of a causal chain of events linking risk factors to a clinical syndrome such as AD. Furthermore, gene expression studies in combination with disease association can nominate transcriptional regulatory mechanisms as a testable culprit for novel AD GWAS loci, such as CLU, ABCA7 and MS4A4A, as discussed. However, beyond gene discovery, the AD GWAS have had an added benefit in that the genotyping they have performed included many thousands of subjects with pertinent intermediate traits, and these genotype data can now be repurposed for discovery studies targeting the intermediate traits [34•, 56•, 100•]. A major limitation of such efforts is the fact that, in many cases, the intermediate phenotypes were not collected systematically in the same manner or using the same strategy across different cohort studies, which complicates the merging of results across individual studies and reduces our statistical power. Nonetheless, such meta-analyses for gene expression, neuropathologic, cognitive and other traits is clearly an important goal for the near future as large sample sizes will be needed for these studies, as has been well demonstrated by the case/control approaches. While repurposing existing data is a valuable activity that will yield insights, we ultimately need to gather as many intermediate traits as possible from the same subjects, as this allows us to fully explore the relationship of these traits and how the effect of a risk factor (genetic or environmental) propagates to influence AD susceptibility. The AD neuroimaging initiative [131] and prospective cohort studies of aging, such as the Mayo Clinic Study of Aging [22], and Religious Order Study and the Memory and Aging Project, which also have brain donations [74], are excellent illustrations of the type of resource that are needed to powerfully investigate the pathophysiology of AD and other neuropsychiatric diseases. Such studies, if they were tenfold larger, would provide ideal platforms for communities of investigators in this field to explore the chain of events linking risk factors to a clinical syndrome. This information will be critical in the successful translation of gene discoveries to viable therapeutic approaches.
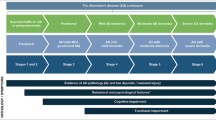
Simplified model for the pathophysiology of AD and its endophenotypes: the flow of major pathogenic mechanisms from top to bottom represent the proposed consequence of events. The interactions between the various pathomechanisms are omitted for simplicity. The dotted arrows and boxes symbolize the pathogenic events that presumably precede the endophenotypes, and the endophenotypes, respectively. The examples of genes that are associated with the endophenotypes are taken from the text. While there are clearly many other plausible endophenotypes in this cascade, only those that are the focus of this review are shown
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–7.
• Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53. This review article summarizes potential sources of heritability for human diseases and traits, which is not explained by GWAS variants, and discusses alternative approaches for discovering the sources for this “missing heritability”.
• Ertekin-Taner N. Gene expression endophenotypes: a novel approach for gene discovery in Alzheimer’s disease. Mol Neurodegener. 2011;6:31. This review article provides a detailed summary of gene expression GWAS studies in human tissues.
Gottesman II, Shields J. Genetic theorizing and schizophrenia. Br J Psychiatry. 1973;122:15–30.
Bearden CE, Freimer NB. Endophenotypes for psychiatric disorders: ready for primetime? Trends Genet. 2006;22:306–13.
Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–45.
Glahn DC, Thompson PM, Blangero J. Neuroimaging endophenotypes: strategies for finding genes influencing brain structure and function. Hum Brain Mapp. 2007;28:488–501.
Ertekin-Taner N, Graff-Radford N, Younkin LH, Eckman C, Baker M, Adamson J, Ronald J, Blangero J, Hutton M, Younkin SG. Linkage of plasma Abeta42 to a quantitative locus on chromosome 10 in late-onset Alzheimer’s disease pedigrees. Science. 2000;290:2303–4.
Gould TD, Gottesman II. Psychiatric endophenotypes and the development of valid animal models. Genes Brain Behav. 2006;5:113–9.
Braak H, Braak E. Neuropathological staging of Alzheimer-related changes. Acta Neuropathol (Berl). 1991;82:239–59.
Ertekin-Taner N. Genetics of Alzheimer’s disease: a centennial review. Neurol Clin. 2007;25:611–67.
Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297:353–6.
Scheuner D, Eckman C, Jensen M, Song X, Citron M, Suzuki N, Bird TD, Hardy J, Hutton M, Kukull W, et al. Secreted amyloid beta-protein similar to that in the senile plaques of Alzheimer’s disease is increased in vivo by the presenilin 1 and 2 and APP mutations linked to familial Alzheimer’s disease. Nat Med. 1996;2:864–70.
Ertekin-Taner N, Graff-Radford N, Younkin LH, Eckman C, Adamson J, Schaid DJ, Blangero J, Hutton M, Younkin SG. Heritability of plasma amyloid beta in typical late-onset Alzheimer’s disease pedigrees. Genet Epidemiol. 2001;21:19–30.
Ertekin-Taner N, Ronald J, Asahara H, Younkin L, Hella M, Jain S, Gnida E, Younkin S, Fadale D, Ohyagi Y, et al. Fine mapping of the alpha-T catenin gene to a quantitative trait locus on chromosome 10 in late-onset Alzheimer’s disease pedigrees. Hum Mol Genet. 2003;12:3133–43.
Ertekin-Taner N, Allen M, Fadale D, Scanlin L, Younkin L, Petersen RC, Graff-Radford N, Younkin SG. Genetic variants in a haplotype block spanning IDE are significantly associated with plasma Abeta42 levels and risk for Alzheimer disease. Hum Mutat. 2004;23:334–42.
Ertekin-Taner N, Ronald J, Feuk L, Prince J, Tucker M, Younkin L, Hella M, Jain S, Hackett A, Scanlin L, et al. Elevated amyloid beta protein (Abeta42) and late onset Alzheimer’s disease are associated with single nucleotide polymorphisms in the urokinase-type plasminogen activator gene. Hum Mol Genet. 2005;14:447–60.
Kauwe JS, Wang J, Mayo K, Morris JC, Fagan AM, Holtzman DM, Goate AM. Alzheimer’s disease risk variants show association with cerebrospinal fluid amyloid beta. Neurogenetics. 2009;10:13–7.
Kauwe JS, Cruchaga C, Mayo K, Fenoglio C, Bertelsen S, Nowotny P, Galimberti D, Scarpini E, Morris JC, Fagan AM, et al. Variation in MAPT is associated with cerebrospinal fluid tau levels in the presence of amyloid-beta deposition. Proc Natl Acad Sci U S A. 2008;105:8050–4.
Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack C, Jagust W, Trojanowski JQ, Toga AW, Beckett L. The Alzheimer’s disease neuroimaging initiative. Neuroimaging Clin N Am. 2005;15:869–77.
Bennett DA, Schneider JA, Buchman AS, Mendes de Leon C, Bienias JL, Wilson RS. The rush memory and aging project: study design and baseline characteristics of the study cohort. Neuroepidemiology. 2005;25:163–75.
Roberts RO, Geda YE, Knopman DS, Cha RH, Pankratz VS, Boeve BF, Ivnik RJ, Tangalos EG, Petersen RC, Rocca WA. The Mayo Clinic Study of Aging: design and sampling, participation, baseline measures and sample characteristics. Neuroepidemiology. 2008;30:58–69.
Glahn DC, Almasy L, Blangero J, Burk GM, Estrada J, Peralta JM, Meyenberg N, Castro MP, Barrett J, Nicolini H, et al. Adjudicating neurocognitive endophenotypes for schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2007;144:242–9.
•• Jack CR Jr, Knopman DS, Jagust WJ, Shaw LM, Aisen PS, Weiner MW, Petersen RC, Trojanowski JQ. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurol. 2010;9:119–28. This review discusses the dynamic change of biomarkers in Alzheimer’s disease and provides a hypothetical model for their longitudinal progression.
Cooper-Knock J, Kirby J, Ferraiuolo L, Heath PR, Rattray M, Shaw PJ. Gene expression profiling in human neurodegenerative disease. Nat Rev Neurol. 2012;8:518–30.
Haroutunian V, Katsel P, Schmeidler J. Transcriptional vulnerability of brain regions in Alzheimer’s disease and dementia. Neurobiol Aging. 2009;30:561–73.
Bossers K, Wirz KT, Meerhoff GF, Essing AH, van Dongen JW, Houba P, Kruse CG, Verhaagen J, Swaab DF. Concerted changes in transcripts in the prefrontal cortex precede neuropathology in Alzheimer’s disease. Brain. 2010;133:3699–723.
Li YJ, Oliveira SA, Xu P, Martin ER, Stenger JE, Scherzer CR, Hauser MA, Scott WK, Small GW, Nance MA, et al. Glutathione S-transferase omega-1 modifies age-at-onset of Alzheimer disease and Parkinson disease. Hum Mol Genet. 2003;12:3259–67.
Capurso C, Panza F, Seripa D, Frisardi V, Imbimbo BP, Verdile G, Vendemiale G, Pilotto A, Solfrizzi V. Polymorphisms in glutathione S-transferase omega-1 gene and increased risk of sporadic Alzheimer disease. Rejuvenation Res. 2010;13:645–52.
Kolsch H, Linnebank M, Lutjohann D, Jessen F, Wullner U, Harbrecht U, Thelen KM, Kreis M, Hentschel F, Schulz A, et al. Polymorphisms in glutathione S-transferase omega-1 and AD, vascular dementia, and stroke. Neurology. 2004;63:2255–60.
Nishimura M, Sakamoto T, Kaji R, Kawakami H. Influence of polymorphisms in the genes for cytokines and glutathione S-transferase omega on sporadic Alzheimer’s disease. Neurosci Lett. 2004;368:140–3.
Ozturk A, Desai PP, Minster RL, Dekosky ST, Kamboh MI. Three SNPs in the GSTO1, GSTO2 and PRSS11 genes on chromosome 10 are not associated with age-at-onset of Alzheimer’s disease. Neurobiol Aging. 2005;26:1161–5.
Wahner AD, Glatt CE, Bronstein JM, Ritz B. Glutathione S-transferase mu, omega, pi, and theta class variants and smoking in Parkinson’s disease. Neurosci Lett. 2007;413:274–8.
• Zou F, Chai HS, Younkin CS, Allen M, Crook J, Pankratz VS, Carrasquillo MM, Rowley CN, Nair AA, Middha S, et al. Brain expression genome-wide association study (eGWAS) identifies human disease-associated variants. PLoS Genet. 2012;8:e1002707. The results of this brain eGWAS suggests that many human disease-associated genetic variants may influence brain transcript levels.
Allen M, Zou F, Chai HS, Younkin CS, Miles R, Nair AA, Crook JE, Pankratz VS, Carrasquillo MM, Rowley CN, et al. Glutathione S-transferase omega genes in Alzheimer and Parkinson disease risk, age-at-diagnosis and brain gene expression: an association study with mechanistic implications. Mol Neurodegener. 2012;7:13.
Webster JA, Gibbs JR, Clarke J, Ray M, Zhang W, Holmans P, Rohrer K, Zhao A, Marlowe L, Kaleem M, et al. Genetic control of human brain transcript expression in Alzheimer disease. Am J Hum Genet. 2009;84:445–58.
Taguchi K, Yamagata HD, Zhong W, Kamino K, Akatsu H, Hata R, Yamamoto T, Kosaka K, Takeda M, Kondo I, et al. Identification of hippocampus-related candidate genes for Alzheimer’s disease. Ann Neurol. 2005;57:585–8.
Chapuis J, Hot D, Hansmannel F, Kerdraon O, Ferreira S, Hubans C, Maurage CA, Huot L, Bensemain F, Laumet G, et al. Transcriptomic and genetic studies identify IL-33 as a candidate gene for Alzheimer’s disease. Mol Psychiatry. 2009;14:1004–16.
•• Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M. Mapping complex disease traits with global gene expression. Nat Rev Genet. 2009;10:184–94. This review discusses the approach of combined eQTL and disease GWA studies to discover regulatory disease loci and pathways.
Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888.
Richards AL, Jones L, Moskvina V, Kirov G, Gejman PV, Levinson DF, Sanders AR, Purcell S, Visscher PM, Craddock N, et al. Schizophrenia susceptibility alleles are enriched for alleles that affect gene expression in adult human brain. Mol Psychiatry. 2012;17:193–201.
Kang HP, Yang X, Chen R, Zhang B, Corona E, Schadt EE, Butte AJ. Integration of disease-specific single nucleotide polymorphisms, expression quantitative trait loci and coexpression networks reveal novel candidate genes for type 2 diabetes. Diabetologia. 2012;55:2205–13.
Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011;43:436–41.
Pittman AM, Fung HC, de Silva R. Untangling the tau gene association with neurodegenerative disorders. Hum Mol Genet. 2006;15 Spec No 2:R188–95.
Grover A, Houlden H, Baker M, Adamson J, Lewis J, Prihar G, Pickering-Brown S, Duff K, Hutton M. 5′ splice site mutations in tau associated with the inherited dementia FTDP-17 affect a stem-loop structure that regulates alternative splicing of exon 10. J Biol Chem. 1999;274:15134–43.
Myers AJ, Pittman AM, Zhao AS, Rohrer K, Kaleem M, Marlowe L, Lees A, Leung D, McKeith IG, Perry RH, et al. The MAPT H1c risk haplotype is associated with increased expression of tau and especially of 4 repeat containing transcripts. Neurobiol Dis. 2007;25:561–70.
Caffrey TM, Joachim C, Wade-Martins R. Haplotype-specific expression of the N-terminal exons 2 and 3 at the human MAPT locus. Neurobiol Aging. 2008;29:1923–9.
Caffrey TM, Joachim C, Paracchini S, Esiri MM, Wade-Martins R. Haplotype-specific expression of exon 10 at the human MAPT locus. Hum Mol Genet. 2006;15:3529–37.
Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–3.
Bu G. Apolipoprotein E and its receptors in Alzheimer’s disease: pathways, pathogenesis and therapy. Nat Rev Neurosci. 2009;10:333–44.
Laws SM, Hone E, Gandy S, Martins RN. Expanding the association between the APOE gene and the risk of Alzheimer’s disease: possible roles for APOE promoter polymorphisms and alterations in APOE transcription. J Neurochem. 2003;84:1215–36.
Bullido MJ, Artiga MJ, Recuero M, Sastre I, Garcia MA, Aldudo J, Lendon C, Han SW, Morris JC, Frank A, et al. A polymorphism in the regulatory region of APOE associated with risk for Alzheimer’s dementia. Nat Genet. 1998;18:69–71.
Wang JC, Kwon JM, Shah P, Morris JC, Goate A. Effect of APOE genotype and promoter polymorphism on risk of Alzheimer’s disease. Neurology. 2000;55:1644–9.
Cramer PE, Cirrito JR, Wesson DW, Lee CY, Karlo JC, Zinn AE, Casali BT, Restivo JL, Goebel WD, James MJ, et al. ApoE-directed therapeutics rapidly clear beta-amyloid and reverse deficits in AD mouse models. Science. 2012;335:1503–6.
Ertekin-Taner N. Genetics of Alzheimer disease in the pre- and post-GWAS era. Alzheimers Res Ther. 2010;2:3.
• Allen M, Zou F, Chai HS, Younkin CS, Crook J, Pankratz VS, Carrasquillo MM, Rowley CN, Nair AA, Middha S, et al. Novel late-onset Alzheimer disease loci variants associate with brain gene expression. Neurology. 2012;79:221–8. This article describes identification of variants at novel LOAD GWAS loci ABCA7, CLU, MS4A4A, which associate with human brain gene expression.
Raj T, Shulman JM, Keenan BT, Chibnik LB, Evans DA, Bennett DA, Stranger BE, De Jager PL. Alzheimer disease susceptibility loci: evidence for a protein network under natural selection. Am J Hum Genet. 2012;90:720–6.
• Ling IF, Bhongsatiern J, Simpson JF, Fardo DW, Estus S. Genetics of clusterin isoform expression and Alzheimer’s disease risk. PLoS One. 2012;7:e33923. This article identifies associations between the protective CLU allele and levels of CLU1 isoform in human brain.
Nuutinen T, Suuronen T, Kauppinen A, Salminen A. Valproic acid stimulates clusterin expression in human astrocytes: implications for Alzheimer’s disease. Neurosci Lett. 2010;475:64–8.
Zhang XZ, Li XJ, Zhang HY. Valproic acid as a promising agent to combat Alzheimer’s disease. Brain Res Bull. 2010;81:3–6.
Tariot PN, Schneider LS, Cummings J, Thomas RG, Raman R, Jakimovich LJ, Loy R, Bartocci B, Fleisher A, Ismail MS, et al. Chronic divalproex sodium to attenuate agitation and clinical progression of Alzheimer disease. Arch Gen Psychiatry. 2011;68:853–61.
Fleisher AS, Truran D, Mai JT, Langbaum JB, Aisen PS, Cummings JL, Jack CR Jr, Weiner MW, Thomas RG, Schneider LS, et al. Chronic divalproex sodium use and brain atrophy in Alzheimer disease. Neurology. 2011;77:1263–71.
Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–93.
Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41:1094–9.
Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, Bis JC, Smith AV, Carassquillo MM, Lambert JC, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–40.
Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43:429–35.
Kukull WA, Larson EB, Reifler BV, Lampe TH, Yerby MS, Hughes JP. The validity of 3 clinical diagnostic criteria for Alzheimer’s disease. Neurology. 1990;40:1364–9.
Galasko D, Hansen LA, Katzman R, Wiederholt W, Masliah E, Terry R, Hill LR, Lessin P, Thal LJ. Clinical-neuropathological correlations in Alzheimer’s disease and related dementias. Arch Neurol. 1994;51:888–95.
Bennett DA, Schneider JA, Aggarwal NT, Arvanitakis Z, Shah RC, Kelly JF, Fox JH, Cochran EJ, Arends D, Treinkman AD, et al. Decision rules guiding the clinical diagnosis of Alzheimer’s disease in two community-based cohort studies compared to standard practice in a clinic-based cohort study. Neuroepidemiology. 2006;27:169–76.
Katzman R, Terry R, DeTeresa R, Brown T, Davies P, Fuld P, Renbing X, Peck A. Clinical, pathological, and neurochemical changes in dementia: a subgroup with preserved mental status and numerous neocortical plaques. Ann Neurol. 1988;23:138–44.
Schmitt FA, Davis DG, Wekstein DR, Smith CD, Ashford JW, Markesbery WR. “Preclinical” AD revisited: neuropathology of cognitively normal older adults. Neurology. 2000;55:370–6.
Galvin JE, Powlishta KK, Wilkins K, McKeel DW Jr, Xiong C, Grant E, Storandt M, Morris JC. Predictors of preclinical Alzheimer disease and dementia: a clinicopathologic study. Arch Neurol. 2005;62:758–65.
Bennett DA, Wilson RS, Boyle PA, Buchman AS, Schneider JA. Relation of neuropathology to cognition in persons without cognitive impairment. Ann Neurol. 2012;72:599–609.
Bennett DA, Schneider JA, Arvanitakis Z, Kelly JF, Aggarwal NT, Shah RC, Wilson RS. Neuropathology of older persons without cognitive impairment from two community-based studies. Neurology. 2006;66:1837–44.
Petersen RC, Parisi JE, Dickson DW, Johnson KA, Knopman DS, Boeve BF, Jicha GA, Ivnik RJ, Smith GE, Tangalos EG, et al. Neuropathologic features of amnestic mild cognitive impairment. Arch Neurol. 2006;63:665–72.
Sonnen JA, Larson EB, Crane PK, Haneuse S, Li G, Schellenberg GD, Craft S, Leverenz JB, Montine TJ. Pathological correlates of dementia in a longitudinal, population-based sample of aging. Ann Neurol. 2007;62:406–13.
Launer LJ, Petrovitch H, Ross GW, Markesbery W, White LR. AD brain pathology: vascular origins? Results from the HAAS autopsy study. Neurobiol Aging. 2008;29:1587–90.
Schneider JA, Arvanitakis Z, Leurgans SE, Bennett DA. The neuropathology of probable Alzheimer disease and mild cognitive impairment. Ann Neurol. 2009;66:200–8.
• Bennett DA, De Jager PL, Leurgans SE, Schneider JA. Neuropathologic intermediate phenotypes enhance association to Alzheimer susceptibility alleles. Neurology. 2009;72:1495–503. This article demonstrates enhanced association of APOE alleles with neuropathology in comparison to disease outcome.
Yu L, Boyle PA, Schneider JA, Segawa E, Wilson RS, Leurgans S, and Bennett DA. APOE ε4 allele is associated with late-life cognitive change through AD pathology. Psychol Aging. (in press).
Mortimer JA, Snowdon DA, Markesbery WR. The effect of APOE-epsilon4 on dementia is mediated by Alzheimer neuropathology. Alzheimer Dis Assoc Disord. 2009;23:152–7.
Stern Y. Cognitive reserve. Neuropsychologia. 2009;47:2015–28.
Honer WG, Barr AM, Sawada K, Thornton AE, Morris MC, Leurgans SE, Schneider JA, Bennett DA. Cognitive reserve, presynaptic proteins and dementia in the elderly. Transl Psychiatry. 2012;2:e114.
Boyle PA, Wilson RS, Schneider JA, Bienias JL, Bennett DA. Processing resources reduce the effect of Alzheimer pathology on other cognitive systems. Neurology. 2008;70:1534–42.
• Kramer PL, Xu H, Woltjer RL, Westaway SK, Clark D, Erten-Lyons D, Kaye JA, Welsh-Bohmer KA, Troncoso JC, Markesbery WR, et al. Alzheimer disease pathology in cognitively healthy elderly: a genome-wide study. Neurobiol Aging. 2011;32:2113–22. This genome-wide study of AD neuropathology in dementia-free subjects identified associations at the RELN locus.
Muller WE, Eckert A, Kurz C, Eckert GP, Leuner K. Mitochondrial dysfunction: common final pathway in brain aging and Alzheimer’s disease–therapeutic aspects. Mol Neurobiol. 2010;41:159–71.
Zilka N, Kazmerova Z, Jadhav S, Neradil P, Madari A, Obetkova D, Bugos O, Novak M. Who fans the flames of Alzheimer’s disease brains? Misfolded tau on the crossroad of neurodegenerative and inflammatory pathways. J Neuroinflamm. 2012;9:47.
Di Paolo G, Kim TW. Linking lipids to Alzheimer’s disease: cholesterol and beyond. Nat Rev Neurosci. 2011;12:284–96.
Muchowski PJ, Wacker JL. Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci. 2005;6:11–22.
Sleegers K, Lambert JC, Bertram L, Cruts M, Amouyel P, Van Broeckhoven C. The pursuit of susceptibility genes for Alzheimer’s disease: progress and prospects. Trends Genet. 2010;26:84–93.
• Chibnik LB, Shulman JM, Leurgans SE, Schneider JA, Wilson RS, Tran D, Aubin C, Buchman AS, Heward CB, Myers AJ, et al. CR1 is associated with amyloid plaque burden and age-related cognitive decline. Ann Neurol. 2011;69:560–9. This article describes the association of a common CR1 variant with cognitive decline, which is mediated by amyloid plaque burden.
Biffi A, Shulman JM, Jagiella JM, Cortellini L, Ayres AM, Schwab K, Brown DL, Silliman SL, Selim M, Worrall BB, et al. Genetic variation at CR1 increases risk of cerebral amyloid angiopathy. Neurology. 2012;78:334–41.
Keenan BT, Shulman JM, Chibnik LB, Raj T, Tran D, Sabuncu MR, Allen AN, Corneveaux JJ, Hardy JA, Huentelman MJ, et al. A coding variant in CR1 interacts with APOE-epsilon4 to influence cognitive decline. Hum Mol Genet. 2012;21:2377–88.
Yu L, Shulman JM, Chibnik L, Leurgans S, Schneider JA, De Jager PL, Bennett DA. The CETP I405 V polymorphism is associated with an increased risk of Alzheimer’s disease. Aging Cell. 2012;11:228–33.
Schlesinger D, Grinberg LT, Alba JG, Naslavsky MS, Licinio L, Farfel JM, Suemoto CK, de Lucena Ferretti RE, Leite RE, de Andrade MP, et al. African ancestry protects against Alzheimer’s disease-related neuropathology. Mol Psychiatry. 2013;18:79–85.
McQueen MB, Bertram L, Lange C, Becker KD, Albert MS, Tanzi RE, Blacker D. Exploring candidate gene associations with neuropsychological performance. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:987–91.
Wilson RS, Leurgans SE, Boyle PA, Bennett DA. Cognitive decline in prodromal Alzheimer disease and mild cognitive impairment. Arch Neurol. 2011;68:351–6.
Diggle P, Heagerty P, Liang K-L, Zeger S. Analysis of longitudinal data. Oxford: Oxford University Press; 2002.
•• Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, Iwatsubo T, Jack CR Jr, Kaye J, Montine TJ, et al. Toward defining the preclinical stages of Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:280–92. These guidelines provide a framework for longitudinal research studies of “preclinical AD” based on the available biomarker, epidemiology and neuropsychology data.
• De Jager PL, Shulman JM, Chibnik LB, Keenan BT, Raj T, Wilson RS, Yu L, Leurgans SE, Tran D, Aubin C, et al. A genome-wide scan for common variants affecting the rate of age-related cognitive decline. Neurobiol Aging. 2012;33:1017. This GWAS of global cognitive decline confirms the expected effect of APOE and identifies a novel locus near PDE7A/MTFR1.
Finkel D, Reynolds CA, Larsson M, Gatz M, Pedersen NL. Both odor identification and ApoE-epsilon4 contribute to normative cognitive aging. Psychol Aging. 2011;26:872–83.
Schiepers OJ, Harris SE, Gow AJ, Pattie A, Brett CE, Starr JM, Deary IJ. APOE E4 status predicts age-related cognitive decline in the ninth decade: longitudinal follow-up of the Lothian Birth Cohort 1921. Mol Psychiatry. 2012;17:315–24.
Knopman DS, Mosley TH, Catellier DJ, Coker LH. Fourteen-year longitudinal study of vascular risk factors, APOE genotype, and cognition: the ARIC MRI Study. Alzheimers Dement. 2009;5:207–14.
Kozauer NA, Mielke MM, Chan GK, Rebok GW, Lyketsos CG. Apolipoprotein E genotype and lifetime cognitive decline. Int Psychogeriatr. 2008;20:109–23.
Seeman TE, Huang MH, Bretsky P, Crimmins E, Launer L, Guralnik JM. Education and APOE-e4 in longitudinal cognitive decline: MacArthur Studies of Successful Aging. J Gerontol B Psychol Sci Soc Sci. 2005;60:P74–83.
Blair CK, Folsom AR, Knopman DS, Bray MS, Mosley TH, Boerwinkle E. APOE genotype and cognitive decline in a middle-aged cohort. Neurology. 2005;64:268–76.
Hu X, Pickering EH, Hall SK, Naik S, Liu YC, Soares H, Katz E, Paciga SA, Liu W, Aisen PS, et al. Genome-wide association study identifies multiple novel loci associated with disease progression in subjects with mild cognitive impairment. Transl Psychiatry. 2011;1:e54.
Elias MF, Beiser A, Wolf PA, Au R, White RF, D’Agostino RB. The preclinical phase of Alzheimer disease: a 22-year prospective study of the Framingham Cohort. Arch Neurol. 2000;57:808–13.
Wilson RS, Aggarwal NT, Barnes LL, Mendes de Leon CF, Hebert LE, Evans DA. Cognitive decline in incident Alzheimer disease in a community population. Neurology. 2010;74:951–5.
Roses AD. Apolipoprotein E affects the rate of Alzheimer disease expression: beta-amyloid burden is a secondary consequence dependent on APOE genotype and duration of disease. J Neuropathol Exp Neurol. 1994;53:429–37.
Blacker D, Haines JL, Rodes L, Terwedow H, Go RC, Harrell LE, Perry RT, Bassett SS, Chase G, Meyers D, et al. ApoE-4 and age at onset of Alzheimer’s disease: the NIMH genetics initiative. Neurology. 1997;48:139–47.
Khachaturian AS, Corcoran CD, Mayer LS, Zandi PP, Breitner JC. Apolipoprotein E epsilon4 count affects age at onset of Alzheimer disease, but not lifetime susceptibility: The Cache County Study. Arch Gen Psychiatry. 2004;61:518–24.
Sando SB, Melquist S, Cannon A, Hutton ML, Sletvold O, Saltvedt I, White LR, Lydersen S, Aasly JO. APOE epsilon 4 lowers age at onset and is a high risk factor for Alzheimer’s disease; a case control study from central Norway. BMC Neurol. 2008;8:9.
Boyle PA, Buchman AS, Wilson RS, Kelly JF, Bennett DA. The APOE epsilon4 allele is associated with incident mild cognitive impairment among community-dwelling older persons. Neuroepidemiology. 2010;34:43–9.
Mayeux R, Small SA, Tang M, Tycko B, Stern Y. Memory performance in healthy elderly without Alzheimer’s disease: effects of time and apolipoprotein-E. Neurobiol Aging. 2001;22:683–9.
Yaffe K, Cauley J, Sands L, Browner W. Apolipoprotein E phenotype and cognitive decline in a prospective study of elderly community women. Arch Neurol. 1997;54:1110–4.
Bondi MW, Salmon DP, Galasko D, Thomas RG, Thal LJ. Neuropsychological function and apolipoprotein E genotype in the preclinical detection of Alzheimer’s disease. Psychol Aging. 1999;14:295–303.
• Caselli RJ, Dueck AC, Osborne D, Sabbagh MN, Connor DJ, Ahern GL, Baxter LC, Rapcsak SZ, Shi J, Woodruff BK, et al. Longitudinal modeling of age-related memory decline and the APOE epsilon4 effect. N Engl J Med. 2009;361:255–63. This longitudinal study on subjects without MCI or dementia showed APOE ε4-related decline in cognition in clinically normal participants.
Bretsky P, Guralnik JM, Launer L, Albert M, Seeman TE. The role of APOE-epsilon4 in longitudinal cognitive decline: MacArthur Studies of Successful Aging. Neurology. 2003;60:1077–81.
Martins CA, Oulhaj A, de Jager CA, Williams JH. APOE alleles predict the rate of cognitive decline in Alzheimer disease: a nonlinear model. Neurology. 2005;65:1888–93.
Bunce D, Fratiglioni L, Small BJ, Winblad B, Backman L. APOE and cognitive decline in preclinical Alzheimer disease and non-demented aging. Neurology. 2004;63:816–21.
Jonker C, Schmand B, Lindeboom J, Havekes LM, Launer LJ. Association between apolipoprotein E epsilon4 and the rate of cognitive decline in community-dwelling elderly individuals with and without dementia. Arch Neurol. 1998;55:1065–9.
Dal Forno G, Rasmusson DX, Brandt J, Carson KA, Brookmeyer R, Troncoso J, Kawas CH. Apolipoprotein E genotype and rate of decline in probable Alzheimer’s disease. Arch Neurol. 1996;53:345–50.
Stern Y, Brandt J, Albert M, Jacobs DM, Liu X, Bell K, Marder K, Sano M, Albert S, Del-Castillo Castenada C, et al. The absence of an apolipoprotein epsilon4 allele is associated with a more aggressive form of Alzheimer’s disease. Ann Neurol. 1997;41:615–20.
Slooter AJ, Houwing-Duistermaat JJ, van Harskamp F, Cruts M, Van Broeckhoven C, Breteler MM, Hofman A, Stijnen T, van Duijn CM. Apolipoprotein E genotype and progression of Alzheimer’s disease: the Rotterdam Study. J Neurol. 1999;246:304–8.
Roses AD. Apolipoprotein E alleles as risk factors in Alzheimer’s disease. Annu Rev Med. 1996;47:387–400.
Petersen RC, Smith GE, Ivnik RJ, Tangalos EG, Schaid DJ, Thibodeau SN, Kokmen E, Waring SC, Kurland LT. Apolipoprotein E status as a predictor of the development of Alzheimer’s disease in memory-impaired individuals. JAMA. 1995;273:1274–8.
Elias-Sonnenschein LS, Viechtbauer W, Ramakers IH, Verhey FR, Visser PJ. Predictive value of APOE-epsilon4 allele for progression from MCI to AD-type dementia: a meta-analysis. J Neurol Neurosurg Psychiatry. 2011;82:1149–56.
Cosentino S, Scarmeas N, Helzner E, Glymour MM, Brandt J, Albert M, Blacker D, Stern Y. APOE epsilon 4 allele predicts faster cognitive decline in mild Alzheimer disease. Neurology. 2008;70:1842–9.
Reitz C, Honig L, Vonsattel JP, Tang MX, Mayeux R. Memory performance is related to amyloid and tau pathology in the hippocampus. J Neurol Neurosurg Psychiatry. 2009;80:715–21.
Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack CR, Jagust W, Trojanowski JQ, Toga AW, Beckett L. Ways toward an early diagnosis in Alzheimer’s disease: the Alzheimer’s Disease Neuroimaging Initiative (ADNI). Alzheimers Dement. 2005;1:55–66.
Acknowledgments
Supported in part by NIA grants P50AG016574, R01AG032990 (NET); R01 AG036836 (PLDJ); P30AG10161, R01AG15819, R01AG17917 (DAB).
Disclosure
None.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Ertekin-Taner, N., De Jager, P.L., Yu, L. et al. Alternative Approaches in Gene Discovery and Characterization in Alzheimer’s Disease. Curr Genet Med Rep 1, 39–51 (2013). https://doi.org/10.1007/s40142-013-0007-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40142-013-0007-5