Abstract
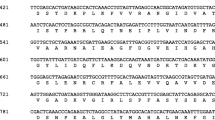
Maize cytosolic glyceraldehyde-3-phosphate dehydrogenase (GAPC) is encoded by a small multi-gene family consisting of gpc1, gpc2, gpc3 and gpc4. GAPC3/4 protein is synthesized in roots during anoxic conditions and is known to be one of the ‘anaerobic polypeptides’. We further analyzed the gpc gene family by isolating full-length cDNA clones of gpc2, gpc3, gpc4 and genomic clones of gpc2 and gpc4. The deduced amino acid sequence of GAPC4 has 99.4% identity with that of GAPC3 as compared to only 81% with either GAPC1 or GAPC2 amino acid sequence. Based on the deduced amino acid sequence identity we designated GAPC1 and GAPC2 as group I (97% identical) and GAPC3 and GAPC4 as group II (99.4% identical). As previously reported for gpc3, transcript levels were also induced for gpc4 by anaerobiosis. Neither heat shock, cold nor salt stress induced the expression of gpc3 or gpc4. In contrast, the transcript accumulation of gpc1 and gpc2 either remained constitutive or decreased in response to anoxia. The upstream regions of gpc2 and gpc4 contain typical eukaryotic promoter features with transcription start points at 76 and 68 bp upstream of their respective translation initiation sites. Transient expression analysis of gpc4 promoter-β-glucuronidase (GUS) reporter gene constructs in bombarded maize suspension culture cells was used to examine the role of 5′-flanking sequence of gpc4. The gpc4 promoter (- 1997 to + 39 bp) was sufficient to induce GUS activity approximately three-fold in response to anaerobiosis. 5′-unidirectional deletion analysis revealed that the critical region of gpc4 required for its induced expression lies between - 290 and - 157. This region has reverse-oriented putative ‘anaerobic response elements’, G-box like sequences, and a GC motif similar to that previously defined as a regulatory element of maize adh1 and Arabidopsis adh, as well as the sequences found in other environmentally inducible genes. The relevance of these elements in conferring anaerobic induction of gpc4 gene expression is discussed.
Similar content being viewed by others
References
Ashburner M, Bonner JJ: The induction of gene activity in Drosophila by heat shock. Cell 17: 241–254 (1979).
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struchl K: Current Protocols in Molecular Biology, pp. 4.8.1–4.8.5. Greene Publishing Associates and Wiley-Interscience, New York (1994).
Bluman AG: Elementary Statistics: A Step by Step Approach, 301 pp. Wm. C. Brown Publishers, Iowa, USA (1992).
Brinkman H, Martinez P, Quigley F, Martin W, Cerff R: Endosymbiotic origin and codon bias of the nuclear gene for chloroplast glyceraldehyde 3-phosphate dehydrogenase from maize. J Mol Evol 26: 320–328 (1987).
Brinkman H, Cerff R, Salmon M, Soll J: Cloning and sequence analysis of cDNAs encoding the cytosolic precursors of subunits GapA and GapB of chloroplast glyceraldehyde-3-phosphate dehydrogenase from pea and spinach. Plant Mol Biol 13: 81–94 (1989).
Chojecki J: Identification and characterization of a cDNA clone for cytosolic glyceraldehyde-3-phosphate dehydrogenase in barley. Carlsberg Res Commun 51: 203–210 (1986).
Christie PJ, Hahn M, Walbot V: Low temperature accumulation of alcohol dehydrogenase-1 mRNA and protein activity in maize and rice seedlings. Plant Physiol 95: 699–706 (1991).
Cone KC, Burr FA, Burr B: Molecular analysis of the maize anthocyanin regulatory locus C1. Proc Natl Acad Sci USA 83: 9631–9635 (1986).
Crespi MD, Zabaleta EJ, Pontis HG, Salerno GL: Sucrose synthase expression during cold acclimation in wheat. Plant Physiol 96: 887–891 (1991).
DeLisle AJ, Ferl RJ: Characterization of the Arabidopsis Adh G-box binding factor. Plant Cell 2: 547–557 (1990).
Dennis ES, Gerlach WL, Pryor AJ, Bennetzen JL, Inglis A, Llewellyn D, Sachs MM, Ferl RJ, Peacock WJ: Molecular analysis of the alcohol dehydrogenase (Adh1) gene of maize. Nucl Acids Res 12: 3983–4000 (1984).
Dennis ES, Sachs MM, Gerlach WL, Finnegan EJ, Peacock WJ: Molecular analysis of the alcohol dehydrogenase 2 (Adh2) gene of maize. Nucl Acids Res 13: 727–743 (1985).
Dennis ES, Gerlach WL, Walker JC, Lavin M, Peacock WJ: Anaerobically regulated aldolase gene of maize: a chimeric origin? J Mol Biol 202: 759–767 (1988).
Dennis ES, Sachs MM, Gerlach WL, Beach L, Peacock WJ: The Ds1 transposable element acts as an intron in the mutant allele Adh1-Fm335 and is spliced from the message. Nucl Acids Res 16: 3815–3828. (1988).
Dewdney J, Conley TR, Shih M-C, Goodman HM: Effects of blue and red light on expression of nuclear genes encoding chloroplast glyceraldehyde 3-phosphate dehydrogenase of Arabidopsis thaliana. Plant Physiol 103: 1115–1121 (1993).
Dolferus R, Jacobs M, Peacock WJ, Dennis ES: Differential interactions of promoter elements in stress responses of the Arabidopsis Adh gene. Plant Physiol 105: 1075–1087 (1994).
Donald RGK, Cashmore AR: Mutation of either G-box or I box sequences profoundly affects expression from the Arabidopsis rbcS-1A promoter. EMBO J 9: 1717–1726 (1990).
Duncan DR, Williams ME, Zehr BE, Widholm JM: The production of callus capable of plant regeneration from immature embryos of numerous Zea mays genotypes. Planta 165: 322–332 (1985).
Fennoy SL, Bailey-Serres J: Post-transcriptional regulation of gene expression in oxygen-deprived roots of maize. Plant J 7: 287–295 (1995).
Foster R, Izawa T, Chua N-H: Plant b ZIP proteins gather at ACGT elements. FASEB J 8: 192–200 (1994).
Gerlach WL, Pryor AJ, Dennis ES, Ferl RJ, Sachs MM, Peacock WJ: cDNA cloning and induction of the alcohol dehydrogenase gene (adh1) of maize. Proc Natl Acad Sci USA 79: 2981–2985 (1982).
Guiltinan MJ, Marcotte WR Jr, Quatrano RS: A plant leucine zipper protein that recognizes an abscisic acid response element. Science 250: 267–271 (1990).
Harris JI, Waters M: Glyceraldehyde 3-phosphate dehydrogenase. In: Boyer PD (ed) The Enzymes, pp. 1–49. Academic Press, New York (1970).
Hake S, Kelley PM, Taylor WC, Freeling M: Coordinate induction of alcohol dehydrogenase 1, aldolase, and other anaerobic RNAs in maize. J Biol Chem 260: 5050–5054 (1985).
Howard EA, Walker JC, Dennis ES, Peacock WJ: Regulated expression of an alcohol dehydrogenase 1 chimeric gene introduced into maize protoplasts. Planta 170: 535–540 (1987).
Jefferson RA, Kavanagh TA, Bevan MW: GUS fusions:β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 (1987).
Kersanach R, Brinkman H, Liaud MF, Zhang DX, Martin W, Cerff R: Five identical intron positions in ancient duplicated genes of eubacterial origin. Nature 367: 387–389 (1994).
Klein TM, Wolf ED, Wu R, Sanford JC: High velocity micro-projectiles for delivering nucleic acids into living cells. Nature 327: 70–73 (1987).
Kohler U, Liaud MF, Mendel RR, Cerff R, Hehl R: The maize GapC4 promoter confers anaerobic reporter gene expression and shows homology to the maize anthocyanin regulatory locus C1. Plant Mol Biol 29: 1293–1298 (1995).
Kohler U, Donath M, Mendel RR, Cerff R, Hehl R: Intronspecific stimulation of anaerobic gene expression and splicing efficiency inmaize cells.Mol Gen Genet 251: 252–258 (1996).
Lal SK, Johnson S, Conway T, Kelley PM: Characterization of a maize cDNA which complements an enolase deficient mutant of Escherichia coli. Plant Mol Biol 16: 787–795 (1991).
Lal SK, Sachs MM: Cloning and characterization of an anaerobically induced cDNA encoding glucose-6-phosphate iso-merase from maize. Plant Physiol 108: 1295–1296 (1995).
Liaud M-F, Valentin C, Martin W, Bouget FY, Kloareg B, Cerff R: The evolutionary origin of red algae as deduced from the nuclear genes encoding cytosolic and chloroplast glyceraldehyde 3-phosphate dehydrogenases from Chondrus crispus. J Mol Evol 38: 319–327 (1994).
Llewellyn DJ, Finnegan EJ, Ellis JG, Dennis ES, Peacock WJ: Structure and expression of an alcohol dehydrogenase 1 gene from Pisumsativum (cv. Green feast). J Mol Biol 195: 115–123 (1987).
Luchrsen KR, de Wet JR, Walbot V: Transient expression analysis in plants using firefly luciferase reporter gene. Meth Enzymol 216: 397 (1992).
Lyons JM, Raison JK: Oxidative activity of mitochondria isolated from plant tissues sensitive and resistant to chilling injury. Plant Physiol 45: 386–389 (1970).
Martinez P, Martin W, Cerff R: Structure, evolution and anaerobic regulation of a nuclear gene encoding cytosolic glyceraldehyde 3-phosphate dehydrogenase genes. J Mol Biol 208: 551–565 (1989).
Nordeen SK: Luciferase reporter gene vectors for analysis of promoters and enhancers. Bio Techniques 6: 454–457 (1988).
Olive MR, Walker JC, Singh K, Dennis ES, Peacock WJ: Functional properties of the anaerobic responsive element of the maize Adh1 gene. Plant Mol Biol 15: 593–604 (1990).
Olive MR, Peacock WJ, Dennis ES: The anaerobic responsive element contains 2 GC-rich sequences essential for binding a nuclear-protein and hypoxic activation of the maize Adh1 promoter. Nucl Acids Res 19: 7053–7060 (1991).
Paul AL, Ferl RJ: Adh1 and Adh2 regulation. Maydica 36: 129–134 (1991).
Peschke VM, Sachs MM: Multiple pyruvate decarboxylase genes in maize are induced by hypoxia. Mol Gen Gent 240: 206–212 (1993).
Peschke VM, Sachs MM: Characterization and expression of transcripts induced by oxygen deprivation in maize (Zea mays L.). Plant Physiol 104: 387–394 (1994).
Rowland LJ, Strommer JN: Anaerobic treatment of maize roots affects transcription of Adh1 and transcript stability. Mol Cell Biol 6: 3368–3372 (1986).
Russell DA, Sachs MM: Differential expression and sequence analysis of the maize glyceraldehyde 3-phosphate dehydrogenase gene family. Plant Cell 1: 793–703 (1989).
Russell DA, Sachs MM: The maize cytosolic glyceraldehyde 3-phosphate dehydrogenase gene family: Organ-specific expression and genetic analysis. Mol Gen Genet 229: 219–228 (1991).
Russell DA, Sachs MM: Protein synthesis in maize during anaerobic and heat stress. Plant Physiol 99: 615–620 (1992).
Sachs MM: Molecular response to anoxic stress in maize. In: Jackson MB, Davies DD, Lambers H (eds) Plant Life under Oxygen Deprivation, pp. 129–139. Academic Publishing, The Hague (1991).
Sachs MM, Freeling M, Okimato R: The anaerobic proteins of maize. Cell 20: 761–767 (1980).
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW: Ribosomal DNA spacer length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci USA 81: 8014–8018 (1984).
Sambrook J, Fritsch EF, Maniatis T: Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY (1989).
Schindler U, Terzagahi W, Beckmann H, Kadesh T, Cashmore AR: DNA binding site preference and transcription activation properties of the Arabidopsis transcription factor GBF1. EMBO J 11: 1275–1289 (1992).
Schulze-Lefert P, Dangl JL, Becker-André M, Hahlbrock K, Schulz W: Inducible in vivo DNA footprints define sequences necessary for UV light activation of parsley chalcone synthase gene. EMBO J 8: 651–656 (1989).
Shen Q, Ho THD: Functional dissection of an abscisic acid (ABA)-inducible gene reveals two independent ABA-responsive complexes each containing a G-box and a novel cis-acting element. Plant Cell 7: 295–307 (1995).
Shih MC, Lazar G, Goodman HM: Evidence in favor of the symbiotic origin of chloroplasts: primary structure and evolution of tobacco glyceraldehyde-3-phosphate dehydrogenases. Cell 47: 73–80 (1986).
Shih M-C, Heinrich P, Goodman HM: Cloning and chromosomal mapping of nuclear genes encoding chloroplast and cytosolic glyceraldehyde 3-phosphate dehydrogenase from Arabidopsis thaliana. Gene 104: 133–138 (1991).
Springer B, Werr W, Starlinger P, Bennett DC, Zokolica M, Freeling M: The shrunken gene on chromosome 9 of Zea mays L. is expressed in various plant tissues and encodes an anaerobic protein. Mol Gen Genet 205: 461–468 (1986).
Subbaiah CC, Zhang J, Sachs MM: Involvement of intracellular calcium in anaerobic gene expression and survival of maize seedlings. Plant Physiol 105: 369–376 (1994).
Subbaiah CC, Bush DS, Sachs MM: Elevation of cytosolic calcium precedes anoxic gene expression in maize suspension-cultured cells. Plant Cell 6: 1747–1762 (1994).
Umeda M, Uchimiya H: Differential transcript levels of genes associated with glycolysis and alcohol fermentation in rice plants (Oryza sativa L.) under submergence stress. Plant Physiol 106: 1015–1022 (1994).
de Vetten NC, Ferl RJ: Characterization of a maize G-box binding factor that is induced by hypoxia. Plant J 7: 589–601 (1995).
Walker JC, Howard EA, Dennis ES, Peacock WJ: DNA sequences required for anaerobic expression of the maize alcohol dehydrogenase 1 gene. Proc Natl Acad Sci USA 84: 6624–6628 (1987).
Wignarajah K, Greenway H: Effect of anaerobiosis on activities of alcohol dehydrogenase and pyruvate decarboxylase in roots of Zea mays. New Phytol 77: 575–584 (1976).
Yang Y, Kwon HB, Peng HP, Shih MC: Stress response and metabolic regulation of glyceraldehyde-3-phosphate dehydrogenase genes in Arabidopsis. Plant Physiol 101: 209–216(1993).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Manjunath, S., Sachs, M.M. Molecular characterization and promoter analysis of the maize cytosolic glyceraldehyde 3-phosphate dehydrogenase gene family and its expression during anoxia. Plant Mol Biol 33, 97–112 (1997). https://doi.org/10.1023/A:1005729112038
Issue Date:
DOI: https://doi.org/10.1023/A:1005729112038