Abstract
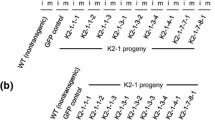
An irregular pattern of transgene silencing was revealed in expression and inheritance studies conducted over multiple generations following transgene introduction by microprojectile bombardment of allohexaploid cultivated oat (Avena sativa L.). Expression of two transgenes, bar and uidA, delivered on the same plasmid was investigated in 23 transgenic oat lines. Twenty-one transgenic lines, each derived from an independently selected transformed tissue culture, showed expression of both bar and uidA while two lines expressed only bar. The relationship of the transgenic phenotypes to the presence of the transgenes in the study was determined using (1) phenotypic scoring combined with Southern blot analyses of progeny, (2) coexpression of the two transgenic phenotypes since the two transgenes always cosegregated, and (3) reactivation of a transgenic phenotype in self-pollinated progenies of transgenic plants that did not exhibit a transgenic phenotype. Transgene silencing was observed in 19 of the 23 transgenic lines and resulted in distorted segregation of transgenic phenotypes in 10 lines. Silencing and inheritance distortions were irregular and unpredictable. They were often reversible in a subsequent generation of self-pollinated progeny and abnormally segregating progenies were as likely to trace back to parents that exhibited normal segregation in a previous generation as to parents showing segregation distortions. Possible causes of the irregular patterns of transgene silencing are discussed.
Similar content being viewed by others
References
Armstrong CL, Parker GB, Pershing JC, Brown SM, Sanders PR, Duncan DR, Stone T, Dean DA, DeBoer DL, Hart J, Howe AR, Morrish FM, Pajeau ME, Petersen WL, Reich BJ, Rodriguez R, Santino CG, Sato SJ, Schuler W, Sims SR, Stehling S, Tarochione LJ, Fromm ME: Field evaluation of European corn borer control in progeny of 173 transgenic corn events expressing an insecticidal protein from Bacillus thuringiensis. Crop Sci 35: 550–557 (1995).
Brandle JE, McHugh SG, James L, Labbe H, Miki BL: Instability of transgene expression in field grown tobacco carrying the csr1–1 gene for sulfonylurea herbicide resistance. Bio/technology 13: 994–998 (1995).
de Carvalho F, Gheysen G, Kushnir S, Van Montagu M, Inzé D, Castresana C: Suppression of β-1,3-glucanase transgene in homozygous plants. EMBO J 11: 2595–2602 (1992).
Christou P: Genetic transformation of crop plants using micro-projectile bombardment. Plant J 2: 275–281 (1992).
Cooley J, Ford T, Christou P: Molecular and genetic characterization of elite transgenic rice plants produced by electric-discharge particle acceleration. Theor Appl Genet 90: 97–104 (1995).
Dehio C, Schell J: Identification of plant genetic loci involved in posttranscriptional mechnism for meiotically reversible transgene silencing. Proc Natl Acad Sci USA 91: 5538–5542 (1994).
Finnegan J, McElroy D: Transgene inactivation: plants fight back! Bio/technology 12: 883–888 (1994).
Fromm ME, Taylor LP, Walbot V: Stable transformation of maize after gene transfer by electroporation. Nature 319: 791–793 (1986).
Fromm ME, Morrish F, Armstrong C, Williams R, Thomas J, Klein TM: Inheritance and expression of chimeric genes in the progeny of transgenic maize plants. Bio/technology 8: 833–839 (1990).
Hosaka K, Kianian SF, McGrath JM, Quiros CF: Development and chromosomal localization of genome-specific DNA markers of Brassica and the evolution of amphidiploids and n=9 species. Genome 33: 131–142 (1990).
Hu J, Quiros CF: Molecular and cytological evidence of deletions in alien chromosomes for two monosomic addition lines of Brassica campestris-oleracea. Theor Appl Genet 81: 221–226 (1991).
Iglesias VA, Moscone EA, Papp I, Neuhuber F, Michalowski S, Phelan T, Spiker S, Matzke M, Matzke AJM: Molecular and cytogenetic analyses of stably and unstably expressed transgene loci in tobacco. Plant Cell 9: 1251–1264 (1997).
Jefferson RA: Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5: 397–405 (1987).
Kumpatla SP, Teng W, Buchholz WG, Hall TC: Epigenetic silencing and 5-azacytidine-mediated reactivation of a complex transgene in rice. Plant Physiol 115: 361–373 (1997).
Matzke MA, Matzke AJM: How and why to plants inactivate homologous (trans)genes? Plant Physiol 107: 679–685 (1995).
McGrath PF, Vincet JR, Lei C-H, Pawlowski WP, Torbert KA, Gu W, Kaeppler HF, Wan Y, Lemaux PG, Rines HW, Somers DA, Larkins BA, Lister RM: Coat protein-mediated resistance to isolates of barley yellow dwarf in oats and barley. Eur J Plant Path 103: 695–710 (1997).
Mittelsten-Scheid O, Jakovleva L, Afsar K, Maluszynska J, Paszkowski J: A change in ploidy can modify epigenetic silencing. Proc Natl Acad Sci USA 93: 7114–7119 (1996).
Nehra NS, Chibbar RN, Leung N, Caswell K, Mallard C, Steinhauer L, Baga M, Kartha KK: Self-fertile transgenic wheat plants regenerated from isolated scutellar tissues following microprojectile bombardment with two distinct gene constructs. Plant J 5: 285–297 (1994).
Pawlowski WP, Somers DA: Transgene inheritance in plants genetically engineered by microprojectile bombardment. Mol Biotechnol 6: 17–30 (1996).
Phillips RL, Kaeppler SM, Olhoft P: Genetic instability of plant tissue cultures: breakdown of normal controls. Proc Natl Acad Sci USA 91: 5222–5226 (1994).
Potrykus I, Paszkowski J, Saul MW, Negrutiu I, amd Shillito RD: Direct gene transfer to plants: facts and future. In: Green CE, Somers DA, Hackett WP, Biesboer DD (eds), Plant Tissue and Cell Culture, pp. 289–302. Alan R. Liss, New York (1987).
Prins M, de Haan P, Luyten R, van Veller M, van Grinsven MQJM, Goldback R: Broad resistance to topoviruses in transgenic tobacco expressing three topoviral nucleoprotein gene sequences. Mol Plant-Microb Interact 8: 85–91 (1995).
Qu R, de Kochko A, Zhang L, Marmey P, Li L, Tiang W, Zhang S, Fauquet CM, Beachy RN: Analysis of a large number of independent transgenic rice plants produced by the biolistic method. In Vitro Cell Devel Biol 32: 233–240 (1996).
Register JC III, Peterson DJ, Bell PJ, Bullock WP, Evans EJ, Frame B, Greenland AJ, Higgs NS, Jepson I, Jiao S, Lewnau CJ, Sillick JM, Wilson HM: Structure and function of selectable and non-selectable transgenes in maize after introduction by particle bombardment. Plant Mol Biol 25: 951–961 (1994).
Rines HW, Luke HH: Selection and regeneration of toxin-insensitive plants from tissue cultures of oat (Avena sativa) susceptible to Helminthosporium victoriae. Theor Appl Genet 71: 16–21 (1985).
Russell DR, Wallace KM, Bathe JH, Martinell BJ, McCabe DE: Stable transformation of Phaseolus vulgaris via electric-discharge mediated particle acceleration. Plant Cell Rep 12: 165–169 (1993).
Somers DA, Rines HW, Gu W, Kaeppler HF, Bushnell WR: Fertile, transgenic oat plants. Bio/technology 10: 1589–1594 (1992).
Southern EM: Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol 98: 503–517 (1975).
Srivastava V, Vasil V, Vasil IK: Molecular characterization of the fate of transgenes in transformed wheat (Triticum aestivum L.). Theor Appl Genet 92: 1031–1037 (1996).
Steel RGD, Torrie JH: Principles and Procedures of Statistics: a Biometrical Approach. McGraw-Hill, New York (1980).
Thompson CJ, Movva NR, Tizard R, Crameri R, Davies JE, Lauwereys M, Botterman J: Characterization of the herbicide-resistance gene bar from Streptomyces hygroscopicus. EMBO J 6: 2519–2523 (1987).
Torbert KA, Rines HW, Somers DA: Use of paromomycin as a selective agent for oat transformation. Plant Cell Rep 14: 635–640 (1995).
Walters DA, Vetsch CS, Potts DE, Lundquist RC: Transformation and inheritance of a hygromycin phosphotransferase gene in maize plants. Plant Mol Biol 18: 189–200 (1992).
Zhang S, Warkentin D, Sun B, Zhong H, Sticklen M: Variation in the inheritance of expression among subclones for unselected (uidA) and selected (bar) transgenes in maize (Zea mays L.). Theor Appl Genet 92: 752–761 (1996).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Pawlowski, W.P., Torbert, K.A., Rines, H.W. et al. Irregular patterns of transgene silencing in allohexaploid oat. Plant Mol Biol 38, 597–607 (1998). https://doi.org/10.1023/A:1006090731414
Issue Date:
DOI: https://doi.org/10.1023/A:1006090731414