Abstract
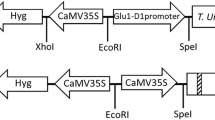
The feasibility of producing plant cell wall polysaccharide-hydrolysing feed enzymes in the endosperm of barley grain was investigated. The coding region of a modified xylanase gene (xynA) from the rumen fungus, Neocallimastix patriciarum, linked with an endosperm-specific promoter from cereal storage protein genes was introduced into barley by Agrobacterium-mediated transformation. Twenty-four independently transformed barley lines with the xylanase gene were produced and analysed. The fungal xylanase was produced in the developing endosperm under the control of either the rice glutelin B-1 (GluB-1) or barley B1 hordein (Hor2-4) promoter. The rice GluB-1 promoter provided an apparently higher expression level of recombinant proteins in barley grain than the barley Hor2-4 promoter in both transient and stable expression experiments. In particular, the mean value for the fungal xylanase activity driven by the GluB-1 promoter in the mature grains of transgenic barley was more than twice that with the Hor2-4 promoter. Expression of the xylanase transgene under these endosperm-specific promoters was not observed in the leaf, stem and root tissues. Accumulation of the fungal xylanase in the developing grains of transgenic barley followed the pattern of storage protein deposition. The xylanase was stably maintained in the grain during grain maturation and desiccation and post-harvest storage. These results indicate that the cereal grain expression system may provide an economic means for large scale production of feed enzymes in the future.
Similar content being viewed by others
References
Bedford MR: Mechanism of action and potential environmental benefits from the use of feed enzymes. Anim Feed Sci Technol 53: 145–155 (1995).
Brandt A, Montembault A, Cameron-Mills V, Rasmussen S: Primary structure of a B1 hordein gene from barley. Carlsberg Res Commun 50: 333–345 (1985).
Bregitzer P, Halbert SE, Lemaux PG: Somaclonal variation in the progeny of transgenic barley. Theo Appl Genet 96: 421–425 (1998).
Cameron-Mills V, Brandt A: A ?-hordein gene. Plant Mol. Biol. 11: 449–461 (1989).
Choct M, Annison G: Anti-nutritive activity of wheat pentosans in broiler diets. Br Poult Sci 31: 811–821 (1990).
Choct M, Hughes RJ, Trimble RP, Angkanaporn K, Annison G: Non-starch polysaccharide-degrading enzymes increase the performance of broiler chickens fed wheat of low apparent metabolizable energy. J Nutr 125, 485–492 (1995).
Classen HL: Enzymes in action. Feed Mix 4: 22–28 (1996).
Demason DA: Endosperm structure and Development. In: Larkins BA, Vasil IK (eds) Cellular and Molecular Biology of Plant Seed Development, pp. 73–115. Kluwer Academic Publishers, Dordrecht, Netherlands (1997).
Denman S, Xue GP, Patel B: Characterization of a Neocallimastix patriciarum cellulase cDNA (celA) homologous to Trichoderma reesei cellobiohydrolase II. App Environ Microbiol 62: 1889–1896 (1996).
Entwistle J: Primary structure of a C-hordein gene from barley. Carlsberg Res Commun 53: 247–258 (1988).
Fincher GB, Stone BA: Cell walls and their components in cereal grain technology. In: Pomeranz Y (ed) Advances in Cereal Science and Technology, Vol. 3, pp. 207–295. American Association of Chemists, Inc., Minnesota (1986).
Finer JJ, Vain P, Jones MW, McMullen MD: Development of the particle inflow gun for DNA delivery to plant cells. Plant Cell Rep 11: 323–328 (1992).
Forde BG, Heyworth A, Pywell J, Kreis M: Nucleotide sequence of a B1 hordein gene and the identification of possible upstream regulatory elements in the endosperm storage protein genes from barley, wheat and maize. Nucl Acids Res 13: 7327–7337 (1985).
Gilbert HJ, Hazlewood GP, Laurie JI, Orpin CG, Xue GP: Homologous catalytic domains in a rumen fungal xylanase: evidence for gene duplication and prokaryotic origin. Mol Microbiol 6: 2065–2072 (1992).
Heim U, Manteuffel R, Bäumlein H, Stenbiss H-H, Wobus U: Transient expression of a lysine-rich vicilin gene of Vicia faba in barley endosperm detected by immunological tissue printing after particle bombardment. Plant Cell Rep 15: 125–128 (1995).
Herbers K, Wilke I, Sonnewald U: A thermostable xylanase from Clostridium thermocellum expressed at high levels in the apoplast of transgenic tobacco has no detrimental effects and is easily purified. Bio/technology 13: 63–66 (1995).
Herbers K, Flint HJ, Sonnewald U: Apoplastic expression of the xylanase and ?(1-3, 1-4) glucanase domains of the xyn D gene from Ruminococcus flavefaciens leads to functional polypeptides in transgenic tobacco plants. Mol Breed 2: 81–87 (1996).
Jefferson RA: Assaying chimeric genes in plants: the Gus gene fusion system. Plant Mol Biol Rep 5: 387–405 (1987).
Knudsen S, Müller M: Transformation of the developing barley endosperm by particle bombardment. Planta 185: 330–336 (1991).
Laliberté J-F, Nicolas O, Durand S, Morosoli R: The xylanase introns from Cryptococcus albidus are accurately spliced in transgenic tobacco plants. Plant Mol Biol 18: 447–451 (1992).
Lazo GR, Stein PA, Ludwig RA: A DNA transformationcompetent Arabidopsis genomic library in Agrobacterium. Bio/technology 9: 963–967 (1991).
Liu JH, Selnger LB, Cheng KJ, Beauchemin KA, Moloney MM: Plant seed oil-bodies as an immobilization matrix for a recombinant xylanase from the rumen fungus Neoxallimastix patriciarum. Mol Breed 3: 463–470 (1997).
Matzke MA, Matzke AJM: How and why do plants inactivate homologous (trans)genes? Plant Physiol 107: 679–685 (1995).
Meyer P: Understanding and controlling transgene expression. Trends Biotechnol 13: 332–337 (1995).
Pettersson D, Aman P: Enzyme supplementation of a poultry diet containing rye and wheat. Br J Nutr 62: 139–149 (1989).
Sambrook J, Fritsch EF, Maniatis T: Molecular Cloning: A Laboratory Manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY (1989).
Shen W-J, Forde BG: Efficient transformation of Agrobacterium spp. by high voltage electroporation. Nucl Acids Res 117: 8385 (1989).
Takaiwa F, Oono K, Kato A: Analysis of the 50 flanking region responsible for the endosperm-specific expression of a rice glutelin chimeric gene in transgenic tobacco. Plant Mol Biol 16: 49–58 (1991).
Takaiwa F, Oono K, Wing D, Kato A: Sequence of three members and expression of a new major subfamily of glutelin genes from rice. Plant Mol Biol 17: 875–885 (1991).
Takaiwa F, Katsube T, Kitagawa S, Hisago T, Kito M, Utsumi S: High level accumulation of soybean glycinin in vacuolederived protein bodies in the endosperm tissue of transgenic tobacco seed. Plant Sci 111: 39–49 (1995).
Thierry D, Vaucheret H: Sequence homology requirements for transcriptional silencing of 35S transgenes and posttranscriptional silencing of nitrite reductase (trans)genes by the tobacco 271 locus. Plant Mol Biol 32: 1075–1083 (1996).
Tingay S, McElroy D, Kalla R, Fieg S, Wang M, Thomton S, Brettell R: Agrobacterium tumefaciens-mediated barley transformation. Plant J 11: 1369–1376 (1997).
Whitelam GC: The production of recombinant proteins in plants. J Sci Food Agric 68: 1–9 (1995).
Witrzens B, Brettell RIS, Murray FR, McElroy D, Li Z, Dennis ES: Comparison of three selectable marker genes for transformation of wheat by microprojectile bombardment. Aust J Plant Physiol 25: 39–44 (1998).
Xue GP, Orpin CG, Gobius KS, Aylward JH, Simpson GD: Cloning and expression of multiple cellulase cDNAs from the anaerobic rumen fungus Neocallimastix patriciarum in Escherichia coli. J Gen Microbiol 138: 1413–1420 (1992).
Xue GP, Denman SE, Glassop D, Johnson JS, Dierens LM, Gobius KS, Aylward JH: Modification of a xylanase cDNA from an anaerobic fungus Neocallimastix patriciarum for high-level expression in Escherichia coli. J Biotechnol 38: 269–277 (1995).
Xue GP, Gobius KS, Ealing PM, Aylward JH: Rumen fungal ?-glucanase and xylanase genes: potential for genetically engineered cereal crops. Proceedings 8th Australian Agronomy Conference 602-605 (1996).
Xue GP, Johnson JS, Smyth DJ, Dierens LM, Wong X, Gobius KS, Aylward JH: Temperature-regulated expression of recombinant proteins from the tac promoter-based expression system in Escherichia coli. Appl Microbiol Biotechnol 45: 120–126 (1996).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Patel, M., Johnson, J.S., Brettell, R.I. et al. Transgenic barley expressing a fungal xylanase gene in the endosperm of the developing grains. Molecular Breeding 6, 113–124 (2000). https://doi.org/10.1023/A:1009640427515
Issue Date:
DOI: https://doi.org/10.1023/A:1009640427515