Abstract
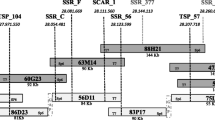
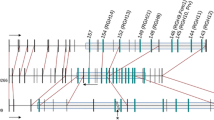
A bacterial artificial chromosome (BAC) library from the dihaploid melon line `PIT92' was constructed with a 6 times coverage of the haploid melon genome. A contig of four BACs around the MRGH63 resistance gene homologue fragment was created. The complete sequence of a 117-kb BAC clone allowed to determine two clearly defined regions, the first one containing a cluster of three resistance gene homologues. Separated by a retrotransposon, that contains large long terminal repeats, the second region presents a group of genes with a conserved distribution in two regions of the Arabidopsis genome. The detailed analysis of this region provides a description of the gene structure and the presence of repetitive sequences in a defined fragment of the genome of Cucumis melo.
Similar content being viewed by others
References
Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W. and Lipman, D.J. 1997. Gapped BLAST and PSIBLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402.
Apweiler, R., Attwood, T.K., Bairoch, A., Bateman, A., Birney, E., Biswas, M., Bucher, P., Cerutti, L., Corpet, F., Croning, M.D.R., et al. 2001. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 29: 37–40.
Arumuganathan, K. and Earle, E.D. 1991. Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9: 208–218.
Bendich, A.J. and Anderson, R.S. 1974. Novel properties of satellite DNA from muskmelon. Proc. Natl. Acad. Sci. USA 71: 1511–1515.
Borodovsky, M. and McIninch, J. 1993. GeneMark: parallel gene recognition for both DNA strands. Computers & Chemistry 17: 123–133.
Burge, C. and Karlin, S. 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78–94.
Casacuberta, E., Puigdomènech, P. and Monfort, A. 2000. Distribution of microsatellites in relation to coding sequences within the Arabidopsis thaliana genome. Plant Sci. 157: 97–104.
Church, G.M. and Gilbert, W. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81: 1991–1995.
Danin-Poleg, Y., Reis, N., Baudracco-Arnas, S., Pitrat, M., Staub, J.E., Oliver, M., ArÚs, P., deVicente, C.M. and Katzir, N. 2000. Simple sequence repeats in Cucumis mapping and map merging. Genome 43: 963–974.
Dixon, M.S., Jones, D.A., Keddie, J.S., Thomas, C.M., Harrison, K. and Jones, J.D. 1996. The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell 84: 451–459.
Durrant, W.E., Rowland, O., Piedras, P., Hammond-Kosack, K.E. and Jones, J.D.G. 2000. cDNA-AFLP reveals a striking overlap in race-specific resistance and wound response gene expression profiles. Plant Cell 12: 963–977.
Emanuelsson, O., Nielsen, H., Brunak, S. and von Heijne, G. 2000. Predicting subcellular localisation of proteins based on their Nterminal amino acid sequence. J. Mol. Biol. 300: 1005–1016.
Feinberg, A.P. and Vogelstein, B. 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132: 6–13.
Frijters, A.C.J., Zhang, Z., van Damme, M., Wang, G.L., Ronald, P.C. and Michelmore, R.W. 1997. Construction of a bacterial artificial chromosome library containing large EcoRI and HindIII genomic fragments of lettuce. Theor. Appl. Genet. 94: 390–399.
Garcia-Mas, J., van Leeuwen, H., Monfort, A., de Vicente, M.C., Puigdomènech, P. and ArÚs, P. 2001. Cloning and mapping of resistance gene homologues in melon. Plant Sci. 161: 165–172.
Gorelick, R.J., Henderson, L.E., Hanser, J.P. and Rein, A. 1988. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: evidence for specific RNA recognition by a 'zinc finger-like’ protein sequence. Proc. Natl. Acad. Sci. USA 85: 8420–8424.
Hamilton, C.M., Frary, A., Xu, Y., Tanksley, S.D. and Zhang, H.B. 1999. Construction of tomato genomic DNA libraries in a binary-BAC (BIBAC) vector. Plant J. 18: 223–229.
Hassanain, H.H., Sharma, Y.K., Moldovan, L., Khramtsov, V., Berliner, L.J., Duvick, J.P. and Goldschmidt-Clermont, P.J. 2000. Plant rac proteins induce superoxide production in mammalian cells. Biochem. Biophys. Res. Commun. 272: 783–788.
Hebsgaard, S.M., Korning, P.G., Tolstrup, N., Engelbrecht, J., Rouze, P. and Brunak, S. 1996. Splice site prediction in Arabidopsis thaliana DNA by combining local and global sequence information. Nucleic Acids Res. 24: 3439–3452.
Jentoft, J.E., Smith, L.M., Fu, X.D., Johnson, M. and Leis, J. 1988. Conserved cysteine and histidine residues of the avian myeloblastosis virus nucleocapsid protein are essential for viral replication but are not 'zinc-binding fingers'. Proc. Natl. Acad. Sci. USA 85: 7094–7098.
Ku, H.M., Vision, T., Liu, J. and Tanksley, S.D. 2000. Comparing sequenced segments of the tomato and Arabidopsis genomes: Large-scale duplication followed by selective gene loss creates a network of synteny. Proc. Natl. Acad. Sci. USA 97: 9121–9126.
Langdon, T., Seago, C., Mende, M., Leggett, M., Thomas, H., Forster, J.W., Thomas, H., Jones, R.N. and Jenkins, G. 2000. Retrotransposition evolution in diverse plant genomes. Genetics 156: 313–325.
Letunic, I., Goodstadt, L., Dickens, N.J., Doerks, T., Schultz, J., Mott, R., Ciccarelli, F., Copley, R.R., Ponting, C.P. and Bork, P. 2002. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 30: 242–244.
Lukashin A. and Borodovsky M. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26: 1107–1115.
Mayer, K., Murphy, G., Tarchini, R., Wambutt, R., Volckaert, G., Pohl, T., Düsterhöft, A., Stiekema, W., Entian, K., Terryn, N. et al. 2001. Conservation ofMicrostructure between a Sequenced Region of the Genome of Rice and Multiple Segments of the Genome of Arabidopsis thaliana. Genome Res. 11: 1167–1174.
Morales, M., Luís-Arteaga, M., Alvarez, J.M., Dolcet-Sanjuan, R., Monfort, A., ArÚs, P. and Garcia-Mas, J. 2002. Marker saturation of the region flanking the gene NSV conferring resistance to the Melon Necrotic Spot Carmovirus (MNSV) in melon. J. Amer. Soc. Hort. Sci. 127: 540–544.
Nicholas, K.B., Nicholas H.B. Jr. and Deerfield, D.W. II. 1997. GeneDoc: Analysis and Visualization of Genetic Variation, EMBNET.NEWS 4: 14, http://www.hgmp.mrc.ac.uk/embnet.news/vol4-2/genedoc.html.
Oliver, M., Garcia-Mas, J., Cardus, M., Pueyo, N., Lopez-Sese, A.L., Arroyo, M., Gomez-Paniagua, H., Arus, P. and de Vicente, M.C. 2001. Construction of a reference linkage map for melon. Genome 44: 836–845.
Parniske, M., Hammond-Kosack, K.E., Golstein, C., Thomas, C.M., Jones, D.A., Harrison, K., Wulff, B.B. and Jones, J.D. 1997. Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell 12: 821–832.
Reese, M.G., Eeckman, F.H., Kulp, D. and Haussler, D. 1997. Improved splice site detection in Genie. Proceedings of the First Annual International Conference on Computational Molecular Biology (RECOMB), Santa Fe, NM, ACM Press, New York.
Rice, P., Longden, I. and Bleasby, A. 2000. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 16: 276–277.
Richter, T.E. and Ronald, P.C. 2000. The evolution of disease resistance genes. Plant Mol. Biol. 42: 195–204.
Rogozin, I.B. and Milanesi, L. 1997. Analysis of donor splice signals in different organisms. J. Mol. Evol. 45: 50–59.
Rossberg, M., Theres, K., Acarkan, A., Herrero, R., Schmitt, T., Schumacher, K., Schmitz, G. and Schmidt, R. 2001. Comparative sequence analysis reveals extensive microcolinearity in the Lateral Suppressor regions of the tomato, Arabidopsis, and Capsella genomes. Plant Cell 13: 979–988.
Salinas-Mondragón, R.E., Garcidueñas-Piña, C. and Guzmán, P. 1999. Early elicitor induction in members of a novel multigene family coding for highly related RING-H2 proteins in Arabidopsis thaliana. Plant Mol. Biol. 40: 579–590.
Salzberg, S.L., Pertea, M., Delcher, A.L., Gardner, M.J. and Tettelin, H. 1999. Interpolated Markov models for eukaryotic gene finding. Genomics 59: 24–31.
Sambrook, J., Fritsch, E.F. and Maniatis, T. 1989. Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
Schmidt, R. 2002. Plant genome evolution: lessons from comparative genomics at the DNA level. Plant Mol. Biol. 48: 21–37.
Smith, R.F., Wiese, B.A., Wojzynski, M.K., Davison, D.B. and Worley, K.C. 1996. BCM Search Launcher–An Integrated Interface to Molecular Biology Data Base Search and Analysis Services Available on the World Wide Web. Genome Res. 6: 454–462.
Smyth, D.R., Kalitsis, P., Joseph, J.L. and Sentry, J.W. 1989. Plant retrotransposon from Lilium henryi is related to Ty3 of yeast and the gypsy group of Drosophila. Proc. Natl. Acad. Sci. USA 86: 5015–5019.
Soltis, P.S., Soltis, D.E. and Chase, M.W. 1999. Angiosperm phylogeny inferred from multiple genes as a tool for comparative biology. Nature 402: 402–404.
Song, W.Y., Pi, L.Y., Wang, G.L., Gardner, J., Holsten, T. and Ronald, P.C. 1997. Evolution of the rice Xa21 disease resistance gene family. Plant Cell 9: 1279–1287.
Sonnhammer, E.L.L., von Heijne, G. and Krogh, A. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proceedings of Sixth Conference on Intelligent Systems for Molecular Biology, Menlo Park, CA, USA, AAAI Press, 175–182.
Staden, R. 1996. The Staden Sequence Analysis Package. Mol. Biotechnol. 5: 233–241.
Strong, S.J., Ohta, Y., Litman, G.W. and Amemiya, C.T. 1997. Marked improvement of PAC and BAC cloning is achieved using electroelution of pulsed-field gel-seperated partial digests of genomic DNA. Nucleic Acids Res. 25: 3959–3961.
Sudupak, M.A., Bennetzen, J.L. and Hulbert, S.H. 1993. Unequal exchange and meiotic instability of disease-resistance genes in the Rp1 region of maize. Genetics 133: 119–125.
Takai, R., Hasegawa, K., Kaku, H., Shibuya, N. and Minami, E. 2001. Isolation and analysis of expression mechanisms of a rice gene, EL5, which shows structural similarity to ATL family from Arabidopsis, in response to N-acetylchitooligosaccharide elicitor. Plant Sci. 160: 577–583.
Tarchini, R., Biddle, P., Wineland, R., Tingey, S. and Rafalski, A. 2000. The complete sequence of 340 kb of DNA around the rice Adh1-Adh2 region reveals interrupted colinearity with maize chromosome 4. Plant Cell 12: 381–391.
Tatusova, T.A. and Madden, T.L. 1999. Blast 2 sequences – a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 174: 247–250.
Temnykh, S., DeClerck, G., Lukashova, A., Lipovich, L., Cartinhour, S. and McCouch, S. 2001. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 11: 1441–1452.
The Arabidopsis Genome Initiative, 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815.
Wang, G.L., Holsten, T.E., Song, W.Y., Wang, H.P. and Ronald, P.C. 1995. Construction of a rice bacterial artificial chromosome library and identification of clones linked to the Xa-21 disease resistance locus. Plant J. 7: 525–533.
Wang, G.L., Ruan, D.L., Song, W.Y., Sideris, S., Chen, L., Pi, L.Y., Zhang, S., Zhang, Z., Fauquet, C., Gaut, B.S., Whalen, M.C. and Ronald, P.C. 1998. Xa21D encodes a receptor-like molecule with a leucine-rich repeat domain that determines race-specific recognition and is subject to adaptive evolution. Plant Cell 10: 765–779.
Woo, S.S., Jiang, J., Gill, B., Paterson, A.H. and Wing, R.A. 1994. Construction and characterisation of a bacterial artificial chromosome library of Sorghum bicolor. Nucleic Acids Res. 22: 4922–4931.
Xu, M., Song, J., Cheng, Z., Jiang, J. and Korban, S.S. 2001. A bacterial artificial chromosome (BAC) library of Malus floribunda 821 and contig construction for positional cloning of the apple scab resistance gene Vf. Genome 44: 1104–1113.
Zhang, H.B., Choi, S., Woo, S.S., Li, Z., and Wing, R.A. 1996. Construction and characterisation of two rice bacterial artificial chromosome libraries from the parents of a permanent recombinant inbred mapping population. Mol. Breed. 2: 11–24.
Zhang, H.B., Zhao, X., Ding, X, Paterson, H. and Wing, R. 1995. Preparation of megabase-size DNA from plant nuclei. Plant J. 7: 175–184.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
van Leeuwen, H., Monfort, A., Zhang, HB. et al. Identification and characterisation of a melon genomic region containing a resistance gene cluster from a constructed BAC library. Microcolinearity between Cucumis melo and Arabidopsis thaliana . Plant Mol Biol 51, 703–718 (2003). https://doi.org/10.1023/A:1022573230486
Issue Date:
DOI: https://doi.org/10.1023/A:1022573230486