Abstract
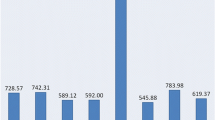
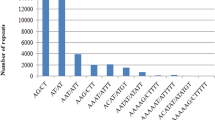
Over 85Mb publicly available expressed sequence tags (ESTs) from four agronomically important crops were used to search for the types and frequencies of simple sequence repeats (SSRs) with motif length of 1-6bp. The frequency of EST-SSRs was one in 11.81kb in rice, 17.42kb in wheat, 23.80kb in soybean, and 28.32kb in maize, respectively. Trinucleotide repeats were the most abundant SSR types with up to 47 trinucleotide repeat units in 100kb ESTs. Compared with dicots, monocots contained GC-rich tri- and hexa- motifs, especially rice where 23.6 CCG motif occurred in 100kb ESTs. 597 EST-SSR primer pairs were designed for wheat, of which, 478 primer pairs had successful PCR amplifications. The percentage of polymorphism was up to 41.8% among wheat varieties, that could be used to construct a transcriptional map of wheat. Cross-species amplification was detected. Among the 478 functional primers from wheat, 255 EST-SSRs amplified fragments in rice, maize and soybean way, indicating high degree of transferability between species, that facilitate comparative mapping and homologous gene cloning.
Similar content being viewed by others
References
Arzimanoglou I.I., Gilbert F. and Barber H.R.K.1998. Microsatellite instability in human solid tumors.Cancer82: 1808–1820.
Bates G. and Lehrach H.1994. Trinucleotide repeat expansions and human genetic disease.BioEssays16: 277–284.
Bennetzen J.L. and Freeling M.1993. Grasses as a single genetic system: Genome composition, colinearity and compatibility.Trends in Genetics9: 259–261.
Buchanan F.C., Adams L.J., Littlejohn R.P., Maddox J.F. and Crawford A.M.1994. Determination of evolutionary relationships among sheep breeds using microsatellites.Genomics22: 397–403.
Cardle L., Ramsay L., Milbourne D., Macaulay M., Marshall D. and Waugh R.2000. Computational and experimental characterization of physically clustered simple sequence repeats in plants.Genetics156: 847–854.
Chen X., Cho Y.G. and McCouch S.R.2002. Microsatellites in Oryza and other plant species.Mol Gen Genomics268: 331–343.
Chin E.C.L., Senior M.L., Shu H. and Smith J.S.C.1996. Maize simple repetitive DNA sequences: abundance and allele variation.Genome39: 866–873.
Fahima T., Röder M.S., Grama A. and Nevo E.1998. Microsatellite DNA polymorphism divergence in Triticum dicoccoides accessions highly resistant to yellow rust.Theor. Appl. Genet.96: 187–195.
Fahima T., Röder M.S., Wendehake K., Kirzhner V.M., Nevo E.2002. Microsatellite polymorphism in natural populations of wild emmer wheat, Triticum dicoccoides, in Israel.Theor. Appl. Genet.104: 17–29.
Fatokun C.A., Menacio-Hautea D.I., Danesh D. and Young N.D.1992. Evidence for orthologous seed weight genes in cowpea and mungbean based upon RFLP mapping.Genetics132: 841–846.
Gale M.D. and Devos K.M.1998. Comparative genetics in the grasses.Proc. Natl. Acad. Sci. USA95: 1971–1974.
Goff S.A., Ricke D., Lan T.H., et al.2002. A draft sequence of the rice genome (Oryza sativa L. ssp. Japonica).Sci.296: 92–100.
Gupta P.K., Balyan H.S., Edwards K.J., Isaac P., Korzun V., Röder M.S., Gautier M.F., Joudrier P., Schlatter A.R., Dubcovsky J., De la Pena R.C., Khairallah M., Penner G., Hayden M.J., Sharp P., Keller B., Wang R.C.C., Hardouin J.P., Jack P. and Leroy P.2002. Genetic mapping of 66 new microsatellite (SSR) loci in bread wheat.Theor. Appl. Genet.105: 413–422.
Hancock J.M.1995. The contribution of slippage-like processes to genome evolution.J. Mol. Evol.41: 1038–1047.
Huang X.Q., Börner A.B., Röder M.S. and Ganal M.W.2002. Assessing genetic diversity of wheat (Triticum aestivum L.) germplasm using microsatellite markers.Theor. Appl. Genet.105: 699–707.
Kantety R.V., La Rota M., Matthews D.E. and Sorrells M.E.2002. Data mining for simple sequence repeats in expressed sequence tags from barley, maize, rice, sorghum and wheat.Plant. Mol. Biol.48: 501–510.
Korzun V., Röder M.S., Ganal M.W., Worland A.J. and Law C.N.1998. Genetic analysis of the dwarfing gene Rht8 in wheat. Part I. Molecular mapping of Rht8 on the short arm of chromosome 2D of bread wheat (Triticum aestivum L.).Theor. Appl. Genet.96: 1104–1109.
Korzun V., Röder M.S., Worland A.J. and Börner A.B. 1997. Intrachromosomal mapping of the genes fro dwarfing (Rht12) and vernalisation response (Vrn1) in wheat by using RFLP and microsatellite markers.Plant Breeding116: 227–232.
Li C.D., Rossnagel B.G. and Scoles G.J.2000. The development of oat microsatellite markers and their use in identifying relationships among Avena species and oat cultivars.Theor. Appl. Genet.101: 1259–1268.
Liu X.M., Smith C.M. and Gill B.S.2002. Identification of microsatellite markers linked to Russian wheat aphid resistance genes Dn4 and Dn6.Theor. Appl. Genet.104: 1042–1048.
Loudet O., Chaillou S., Camilleri C., Bouchez D. and Daniel-Vedele F.2002. Bay-0 × Shahdara recombinant inbred line population: a powerful tool for the genetic dissection of complex traits in Arabidopsis.Theor. Appl. Genet.104: 1173–1184.
Ma X.F., Wanous M.K., Houchins K., Rodrguez mila M.A., Goicoechea P.G., Wang Z., Xie M., Gustafson J.P.2001. Molecular linkage mapping in rye (Secale cereale L.).Theor. Appl. Genet.102: 517–523.
Matsuoka Y., Mitchell S.E., Kresovich S., Goodman M. and Doebley J.2002. Microsatellite in Zea-variability, patterns of mutations, and use for evolutionary studies.Theor. Appl. Genet.104: 436–450.
Morgante M., Hanafey M. and Powell W.2002. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes.Nat. Genet.30: 194–200.
Peng J.H., Fahima T., Röder M.S., Li Y.C., Dahan A., Grama A., Ronin Y.I., Korol A.B. and Nevo E.1999. Microsatellite tagging of the stripe-rust resistance gene YrH52 derived from wild emmer wheat, Triticum dicoccoides, and suggestive negative crossover interference on chromosome 1B.Theor. Appl. Genet.98: 862–872.
Reddy P.S. and Housman D.E.1997. The complex pathology of trinucleotide repeats.Curr. Opin. Cell. Biol.9: 364–372.
Röder M.S., Korzun V., Wendehake K., Plaschke J., Tixier M.H., Leroy P. and Ganal M.W.1998. A microsatellite map of wheat.Genetics149: 2007–2023.
Stack S., Campbell L., Henderson K., Eujayl I., Hanafey M., Powell W. and Wolters P.2000. Development of EST-derived microsatellite markers for mapping and germplasm analysis in wheat.Plant & Animal Genome conference, San Diego, California, USA. pp. 102.
Stephenson P., Bryan G., Kirby J., Collins A., Devos K., Busso C. and Gale M.1998. Fifty new microsatellite loci for the wheat genetic map.Theor. Appl. Genet.97: 946–949.
Temnykh S., DeClerck G., Lukashova A., Lipovich L., Cartinhour S. and McCouch S.2001. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon association, and genetic marker potential.Genome Res.11: 1441–1452.
Tóth G., Gáspári Z. and Jurka J.2000. Microsatellites in different eukaryotic genomes: Survey and analysis.Genome Res10: 967–981.
Warren S.T. and Nelson D.L.1993. Trinucleotide repeat expansions in neurological disease.Curr. Opin. Neurobiol.3: 757–759.
Wooster R., Cleton-Jansen A.M., Collins N., Mangion J., Cornelis R.S., Cooper C.S., Gusterson B.A., Ponder B.A.J., von Deimling A., Wiestler O.D., et al.1994. Instability of short tandem repeats (microsatellites) in human cancer.Nat. Genet6: 152–156.
Xie C.J., Sun Q.X., Ni Z.F., Yang T.M., Nevo E. and Fahima T.2002. Chromosomal location of a Triticum dicoccoides-derived powdery mildew resistance gene in common wheat by using microsatellite markers.Theor. Appl. Genet.104: 1164–1172.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Gao, L., Tang, J., Li, H. et al. Analysis of microsatellites in major crops assessed by computational and experimental approaches. Molecular Breeding 12, 245–261 (2003). https://doi.org/10.1023/A:1026346121217
Issue Date:
DOI: https://doi.org/10.1023/A:1026346121217