Abstract
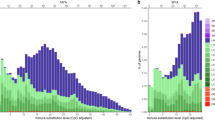
Retrotransposons, transposable elements related to animal retroviruses, are found in all eukaryotes investigated and make up the majority of many plant genomes1,2,3,4,5. Their ubiquity points to their importance, especially in their contribution to the large-scale structure of complex genomes. The nature and frequency of retro-element appearance, activation and amplification are poorly understood in all higher eukaryotes. Here we employ a novel approach to determine the insertion dates for 17 of 23 retrotransposons found near the maize adh1 gene, and two others from unlinked sites in the maize genome, by comparison of long terminal repeat (LTR) divergences with the sequence divergence between adh1 in maize and sorghum. All retrotransposons examined have inserted within the last six million years, most in the last three million years. The structure of the adh1 region appears to be standard relative to the other gene-containing regions of the maize genome, thus suggesting that retrotransposon insertions have increased the size of the maize genome from approximately 1200 Mb to 2400 Mb in the last three million years. Furthermore, the results indicate an increased mutation rate in retrotransposons compared with genes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Flavell, A.J. et al. Ty1-copia group retrotransposons are ubiquitous and heterogeneous in higher plants. Nucleic Acids Res. 20, 3639–3644 (1992).
Grandbastien, M.A. Retroelements in higher plants. Trends Genet. 8, 103–108 (1992).
Voytas, D.F. et al. copia-like retrotransposons are ubiquitous among plants. Proc. Natl Acad. Sci. USA 89, 7124– 7128 (1992).
Bennetzen, J.L. The contributions of retroelements to plant genome organization, function and evolution. Trends Microbiol. 4, 347– 353 (1996).
SanMiguel, P. et al. Nested retrotransposons in the intergenic regions of the maize genome. Science 274, 765– 768 (1996).
Lewin B. Genes VI. 603–604 (Oxford University Press, New York, 1997).
Gaut, B.S., Morton, B.R., McCaig, B.C. & Clegg, M.T. Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc. Natl Acad. Sci. USA 93, 10274–10279 (1996).
Gaut, B.S. & Doebley, J.F. DNA sequence evidence for the segmental allotetraploid origin of maize. Proc. Natl Acad. Sci. USA 94, 6809–6814 ( 1997).
Johns, M.A., Strommer, J.N. & Freeling, M. Exceptionally high levels of restriction site polymorphism in DNA near the maize Adh1 gene. Genetics 105 , 733–743 (1983).
Avramova, Z. et al. Gene identification in a complex chromosomal continuum by local genomic cross-referencing. Plant J. 10, 1163–1168 (1996).
Dooner, H.K. & Martinezferez, I.M. Recombination occurs uniformly within the bronze gene, a meiotic recombination hotspot in the maize genome. Plant Cell 9, 1633– 1646 (1997).
Gaut, B.S. & Clegg, M.T. Molecular evolution of the Adh1 locus in the genus Zea. Proc. Natl Acad. Sci. USA. 90, 5095–5099 ( 1993).
Hu, W.S. & Temin, H.M. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudodiploidy and high rate of genetic recombination. Proc. Natl Acad. Sci. USA 87 , 1556–1560 (1990).
Bennetzen, J.L. et al. Active maize genes are unmodified and flanked by diverse classes of modified, highly repetitive DNA. Genome 37, 565–576 (1994).
Coulondre, C., Miller, J.H., Farabaugh, P.J. & Gilbert, W. Molecular basis of base substitution hotspots in Escherichia coli. Nature 274, 775–780 (1978).
Gruenbaum, Y., Navey-Many, T., Cedar, H. & Razin, A. Sequence specificity of methylation in higher plant DNA. Nature 292, 860–862 ( 1981).
Selker, E.U. & Stevens, J.N. DNA methylation at asymmetric sites is associated with numerous transition mutations. Proc. Natl Acad. Sci. USA 82, 8114–8118 (1985).
Yoder, J.A. & Bestor, T.H. Genetic analysis of genomic methylation patterns in plants and mammals. Biol. Chem. 377, 605–610 (1996).
Jin, Y.K. & Bennetzen, J.L. Structure and coding properties of Bs1, a maize retrovirus-like transposon. Proc. Natl Acad. Sci. USA 86, 6235–6239 ( 1989).
White, S.E., Habera, L.F. & Wessler, S.R. Retrotransposons in the flanking regions of normal plant genes: a role for copia-like elements in the evolution of gene structure and expression. Proc. Natl Acad. Sci. USA 91, 11792–11796 (1994).
Turcich, M.P. et al. PREM-2, a copia-type retroelement in maize is expressed preferentially in early microspores. Sex. Plant Reprod. 9, 65–74 (1996).
Hu, W., Das, O.P. & Messing, J. Zeon-1, a member of a new maize retrotransposon family. Mol. Gen. Genet. 248, 471– 480 (1995).
Flavell, R.B., Bennett, M.D., Smith, J.B. & Smith, D.B. Genome size and the proportion of repeated nucleotide sequence DNA in plants. Biochemical Genetics 12, 257– 269 (1974).
Devereux, J., Haeberli, P. & Smithies, O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12, 387– 395 (1984).
Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120 (1980).
Yang, Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13, 555–556 (1997).
Kumar, S., Tamura, K. & Nei, M. MEGA: Molecular Evolutionary Genetics Analysis software for microcomputers. Comput. Appl. Biosci. 10, 189– 191 (1994).
Nei, M. & Gojobori, T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3, 418–426 (1986).
Gaut, B.S. & Weir, B.S. Detecting substitution-rate heterogeneity among regions of a nucleotide sequence. Mol. Biol. Evol. 11, 620–629 (1994).
Sokal, R.R. & Rohlf, F.J. Biometry. (Freeman, New York, 1995).
Acknowledgements
We thank S. Frank and S. Subramanian for technical assistance and Z. Avramova for useful discussions. This work was supported by grants from the USDA to J.L.B. and B.S.G.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
SanMiguel, P., Gaut, B., Tikhonov, A. et al. The paleontology of intergene retrotransposons of maize. Nat Genet 20, 43–45 (1998). https://doi.org/10.1038/1695
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/1695
This article is cited by
-
Evolutionary dynamics of the LTR-retrotransposon crapaud in the Podospora anserina species complex and the interaction with repeat-induced point mutations
Mobile DNA (2024)
-
The role of transposon inverted repeats in balancing drought tolerance and yield-related traits in maize
Nature Biotechnology (2023)
-
The genome of the glasshouse plant noble rhubarb (Rheum nobile) provides a window into alpine adaptation
Communications Biology (2023)
-
Repeated turnovers keep sex chromosomes young in willows
Genome Biology (2022)
-
Comparative analysis of transposable elements provides insights into genome evolution in the genus Camelus
BMC Genomics (2021)