Abstract
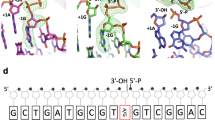
Non-coding apurinic/apyrimidinic (AP) sites in DNA are continually created in cells both spontaneously and by damage-specific DNA glycosylases1. The biologically critical human base excision repair enzyme APE1 cleaves the DNA sugar-phosphate backbone at a position 5′ of AP sites to prime DNA repair synthesis2,3,4. Here we report three co-crystal structures of human APE1 bound to abasic DNA which show that APE1 uses a rigid, pre-formed, positively charged surface to kink the DNA helix and engulf the AP-DNA strand. APE1 inserts loops into both the DNA major and minor grooves and binds a flipped-out AP site in a pocket that excludes DNA bases and racemized β-anomer AP sites. Both the APE1 active-site geometry and a complex with cleaved AP-DNA and Mn2+ support a testable structure-based catalytic mechanism. Alanine substitutions of the residues that penetrate the DNA helix unexpectedly show that human APE1 is structurally optimized to retain the cleaved DNA product. These structural and mutational results show how APE1 probably displaces bound glycosylases and retains the nicked DNA product, suggesting that APE1 acts in vivo to coordinate the orderly transfer of unstable DNA damage intermediates between the excision and synthesis steps of DNA repair.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lindahl,, T. Instability and decay of the primary structure of DNA. Nature 362, 709–715 (1993).
Wilson, D. M. & Thompson, L. H. Life without DNA repair. Proc. Natl Acad. Sci. USA 94, 12754– 12757 (1997).
Wilson, D. M., Takeshita, M., Grollman, A. P. & Demple, B. Incision activity of human apurinic endonuclease (Ape) at abasic site analogs in DNA. J. Biol. Chem. 270, 16002– 16007 (1995).
Izumi, T. & Mitra, S. Deletion analysis of human AP-endonuclease: minimum sequence required for the endonuclease activity. Carcinogenesis 19, 525–527 ( 1998).
Lindahl, T. & Wood, R. D. Quality control in DNA repair. Science 286, 1897–1905 ( 1999).
Tsutakawa, S. E., Jingami, H. & Morikawa, K. Recognition of a TG mismatch: the crystal structure of very short patch repair endonuclease in complex with a DNA duplex. Cell 99, 615–623 ( 1999).
Hosfield, D. J., Guan, Y., Haas, B. J., Cunningham, R. P. & Tainer, J. A. Structure of the DNA repair enzyme endonuclease IV and its DNA complex: double-nucleotide flipping at abasic sites and three-metal-ion catalysis. Cell 98, 397– 408 (1999).
Gorman, M. A. et al. The crystal structure of the human DNA repair endonuclease HAP1 suggests the recognition of extra-helical deoxyribose at DNA abasic sites. EMBO J. 16, 6548–6558 (1997).
Wilson, D. M., Takeshita, M. & Demple, B. Abasic site binding by the human apurinic endonuclease, Ape, and determination of the DNA contact sites. Nucleic Acids Res. 25, 933–939 ( 1997).
Withka, J. M., Wilde, J. A. & Bolton, P. H. Characterization of conformational features of DNA heteroduplexes containing aldehydic abasic sites. Biochemistry 30, 9931–9940 ( 1991).
Mol, C. D., Kuo, C. –F., Thayer, M. M., Cunningham, R. P. & Tainer, J. A. Structure and function of the multifunctional DNA-repair enzyme exonuclease III. Nature 374, 381– 386 (1995).
Erzberger, J. P. & Wilson, D. M. The role of Mg2+ and specific amino acid residues in the catalytic reaction of the major human abasic endonuclease: new insights from EDTA-resistant incision of acyclic abasic site analogs and site-directed mutagenesis. J. Mol. Biol. 290, 447–457 (1999).
Izumi, T. et al. Intragenic suppression of an active site mutation in the human apurinic/apyrimidinic endonuclease. J. Mol. Biol. 287 , 47–57 (1999).
Kane, C. M. & Linn, S. Purification and characterization of an apurinic/apyrimidinic endonuclease from HeLa cells. J. Biol. Chem. 256, 3405–3414 ( 1981).
Masuda, Y., Bennett, R. A. & Demple, B. Dynamics of the interaction of human apurinic endonuclease (Ape1) with its substrate and product. J. Biol. Chem. 273, 30352–30359 (1998).
Bennett, R. A. O., Wilson, D. M., Wong, D. & Demple, B. Interaction of human apurinic endonuclease and DNA polymerase β in the base excision repair pathway. Proc. Natl Acad. Sci. USA 94, 7166–7169 (1997).
Prasad, R. et al. Specific interaction of DNA polymerase β and DNA ligase I in a multiprotein base excision repair complex from bovine testis. J. Biol. Chem. 271, 16000–16007 (1996).
Parikh, S. S. et al. Base–excision repair initiation revealed by crystal structures and DNA–binding kinetics of human uracil–DNA glycosylase bound to DNA. EMBO J. 17, 5214– 5226 (1998).
Waters, T. R., Gallinari, P., Jiricny, J. & Swann, P. F. Human thymine DNA glycosylase binds to apurinic sites in DNA but is displaced by human apurinic endonuclease 1. J. Biol. Chem. 274 , 67–74 (1999).
Chen, D. S., Herman, V. & Demple, B. Two distinct human DNA diesterases that hydrolyze 3′–blocking deoxyribose fragments from oxidized DNA. Nucleic Acids Res. 19, 5907–5914 (1991).
Horton, J. K., Srivastava, D. K., Zmudzka, B. Z. & Wilson, S. H. Strategic down–regulation of DNA polymerase beta by antisense RNA sensitizes mammalian cells to specific DNA damaging agents. Nucleic Acids Res. 23, 3810–3815 ( 1995).
Kubota, Y. et al. Reconstitution of DNA base excision–repair with purified human proteins: interaction between DNA polymerase β and the XRCC1 protein. EMBO J. 15, 6662–6670 (1996).
Caldecott, K. W., McKeown, C. K., Tucker, J. D., Ljungquist, S. & Thompson, L. H. An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol. Cell. Biol. 14, 68–76 (1994).
Takeshita, M., Chang, C. N., Johnson, F., Will, S. & Grollman, A. P. Oligodeoxynucleotides containing synthetic abasic sites. Model substrates for DNA polymerases and apurinic/apyrimidinic endonucleases. J. Biol. Chem. 262, 10171 –10179 (1987).
Erzberger, J. P., Barsky, D., Scharer, O. D., Colvin, M. E. & Wilson, D. M. Elements in abasic site recognition by the major human and Escherichia coli apurinic/apyrimidinic endonucleases. Nucleic Acids Res. 26, 2771– 2778 (1998).
Otwinowski,, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–325 ( 1997).
Navaza, J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. A 50, 157–163 ( 1994).
Brünger, A. T., Kuriyan, J. & Karplus, M. Crystallographic R factor refinement by molecular dynamics. Science 235, 458–460 (1987).
Read, R. J. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr. A 42, 140 –149 (1986).
McRee, D. E. XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125, 156–165 (1999).
Acknowledgements
We thank S. S. Parikh, C. D. Putnam, D. S. Daniels and D. J. Hosfield for helpful discussions, and the staff and facilities at SSRL. This work was supported by the NIH, by a Laboratory Directed Research and Development grant from the Lawrence Berkeley Laboratory, by a cancer research supplement administered through LBNL from the National Cancer Institute (to J.A.T., S.M. and P. Cooper), the Skaggs Institute for Chemical Biology and a Special Fellowship from the Leukemia Society of America (to C.D.M.).
Author information
Authors and Affiliations
Corresponding author
Supplementary information
41586_2000_BF35000249_MOESM1_ESM.gif
Figure 1: DNA-bound AP Endonuclease (APE1) structures and mutants reveal abasic DNA binding to coordinate DNA repair (animated version) (GIF 954 kb)
Rights and permissions
About this article
Cite this article
Mol, C., Izumi, T., Mitra, S. et al. DNA-bound structures and mutants reveal abasic DNA binding by APE1 DNA repair and coordination. Nature 403, 451–456 (2000). https://doi.org/10.1038/35000249
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35000249
This article is cited by
-
Covalent PARylation of DNA base excision repair proteins regulates DNA demethylation
Nature Communications (2024)
-
Regulation of base excision repair during adipogenesis and osteogenesis of bone marrow-derived mesenchymal stem cells
Scientific Reports (2023)
-
Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome
Cell Discovery (2023)
-
Accumulation of DNA damage alters microRNA gene transcription in Arabidopsis thaliana
BMC Plant Biology (2022)
-
Progress in cancer drug delivery based on AS1411 oriented nanomaterials
Journal of Nanobiotechnology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.