Abstract
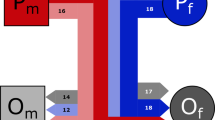
In 1947, it was suggested that, in humans, the mutation rate is dramatically higher in the male germ line than in the female germ line1. This hypothesis has been supported by the observation that, among primates, Y-linked genes evolved more rapidly than homologous X-linked genes2,3,4,5,6. Based on these evolutionary studies, the ratio (αm) of male to female mutation rates in primates was estimated to be about 5. However, selection could have skewed sequence evolution in introns and exons7,8,9,10. In addition, some of the X–Y gene pairs studied lie within chromosomal regions with substantially divergent nucleotide sequences7,11,12. Here we directly compare human X and Y sequences within a large region with no known genes. Here the two chromosomes are 99% identical, and X–Y divergence began only three or four million years ago, during hominid evolution13,14,15. In apes, homologous sequences exist only on the X chromosome. We sequenced and compared 38.6 kb of this region from human X, human Y, chimpanzee X and gorilla X chromosomes. We calculated αm to be 1.7 (95% confidence interval 1.15–2.87), significantly lower than previous estimates in primates. We infer that, in humans and their immediate ancestors, male and female mutation rates were far more similar than previously supposed.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
References
Haldane, J. B. S. The mutation rate of the gene for haemophilia, and its segregation ratios in males and females. Ann. Eugen. 13, 262 –271 (1947).
Miyata, T., Hayashida, H., Kuma, K., Mitsuyasu, K. & Yasunaga, T. Male-driven molecular evolution: A model and nucleotide sequence analysis. Cold Spring Harbor Symp. Quant. Biol. 52, 863–867 (1987).
Shimmin, L. C., Chang, B. H. -J. & Li, W. -H. Male-driven evolution of DNA sequences. Nature 362, 745–747 ( 1993).
Shimmin, L. C., Chang, B. H. -J. & Li, W. -H. Contrasting rates of nucleotide substitution in the X-linked and Y-linked zinc finger genes. J. Mol. Evol. 39, 569–578 (1994).
Chang, B. H.-J., Hewett-Emmett, D. & Li, W.-H. Male-to-female ratios of mutation rate in higher primates estimated from intron sequences. Zool. Stud. 35, 36–48 (1996).
Huang, W., Chang, B. H.-J., Gu, X., Hewett-Emmett, D. & Li, W.-H. Sex differences in mutation rate in higher primates estimated from AMG intron sequences. J. Mol. Evol. 44, 463–465 (1997).
Shimmin, L. C., Chang, B. H.-J., Hewett-Emmett, D. & Li, W.-H. Potential problems in estimating the male-to-female mutation rate ratio from DNA sequence data. J. Mol. Evol. 37, 160 –166 (1993).
Charlesworth, B. The effect of background selection against deleterious alleles on weakly selected, linked variants. Genet. Res. 63, 213– 227 (1994).
Li, W.-H. Molecular Evolution (Sinauer, Sunderland, 1997).
McVean, G. T. & Hurst, L. D. Evidence for a selectively favourable reduction in the mutation rate of the X chromosome. Nature 386, 388–392 (1997).
Bulmer, M. Neighboring base effects on substitution rates in pseudogenes. Mol. Biol. Evol. 3, 322–329 (1986).
Wolfe, K. H., Sharp, P. M. & Li, W.-H. Mutation rates differ among regions of the mammalian genome. Nature 337, 283–285 (1989).
Page, D. C., Harper, M. E., Love, J. & Botstein, D. Occurrence of a transposition from the X-chromosome long arm to the Y-chromosome short arm during human evolution. Nature 311, 119– 123 (1984).
Mumm, S., Molini, B., Terrell, J., Srivastava, A. & Schlessinger, D. Evolutionary features of the 4-Mb Xq21. 3 XY homology region revealed by a map at 60-kb resolution. Genome Res. 7, 307–314 (1997).
Schwartz, A. et al. Reconstructing hominid Y evolution: X-homologous block, created by X-Y transposition, was disrupted by Yp inversion through LINE-LINE recombination. Hum. Mol. Genet. 7, 1– 11 (1998).
Tajima, F. & Nei, M. Estimation of evolutionary distance between nucleotide sequences. Mol. Biol. Evol. 1, 269–285 (1984).
Kaessmann, H., Heissig, F., von Haeseler, A. & Paabo, S. DNA sequence variation in a non-coding region of low recombination on the human X chromosome. Nature Genet. 22, 78 –81 (1999).
Kaessmann, H., Wiebe, V. & Paabo, S. Extensive nuclear DNA sequence diversity among chimpanzees. Science 286, 1159–1162 (1999).
Ellegren, H. & Fridolfsson, A. K. Male-driven evolution of DNA sequences in birds. Nature Genet. 17, 182–184 (1997).
Vogel, F. & Motulsky, A. G. Human Genetics (Springer, Berlin, 1997).
Crow, J. The high spontaneous mutation rate: Is it a health risk? Proc. Natl Acad. Sci. USA 94, 8380–8386 (1997).
Wilkin, D. J. et al. Mutations in fibroblast growth-factor receptor 3 in sporadic cases of achondroplasia occur exclusively on the paternally derived chromosome. Am. J. Hum. Genet. 63, 711– 716 (1998).
Shizuya, H. B. et al. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc. Natl Acad. Sci. USA 89, 8794– 8797 (1992).
Burge, C. & Karlin, S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268, 78–94 (1997).
Uberbacher, E. C. & Mural, R. J. Locating protein-coding regions in human DNA sequences by a multiple sensor-neural network approach. Proc. Natl Acad. Sci. USA 88, 11261– 11265 (1991).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Rozen, S. & Skaletsky, H. in Bioinformatics Methods and Protocols (eds Misener, S. & Krawetz, S. A.) (Humana, Totowa, 1999).
Agresti, A. Categorical Data Analysis (Wiley, New York, 1990).
Li, W. -H. A statistical test of phylogenies estimated from sequence data. Mol. Biol. Evol. 6, 424–435 (1989).
Bishop, Y. V. V., Feinberg, S. E. & Holland, P. W. Discrete Multivariate Analysis (MIT Press, Cambridge, Massachusetts, 1975).
Acknowledgements
We thank A. Schwartz for identifying homologous X and Y-chromosomal BACs, colleagues at the Whitehead Institute/MIT Centre for Genome Research for sequencing those BACs, and J. Bradley, A. Chakravarti, B. Charlesworth, A. Clark, D. Haig, T. Kawaguchi, L. Kruglyak, F. Lewitter, Y.-F. Lim, D. Reich, W. Rice, S. Rozen, C. Tilford and J. Wang for comments on the manuscript. Supported in part by the NIH.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bohossian, H., Skaletsky, H. & Page, D. Unexpectedly similar rates of nucleotide substitution found in male and female hominids. Nature 406, 622–625 (2000). https://doi.org/10.1038/35020557
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35020557
This article is cited by
-
Strict evolutionary conservation followed rapid gene loss on human and rhesus Y chromosomes
Nature (2012)
-
Variation in genome-wide mutation rates within and between human families
Nature Genetics (2011)
-
Understanding Neutral Genomic Molecular Clocks
Evolutionary Biology (2007)
-
Substitution Rate Heterogeneity and the Male Mutation Bias
Journal of Molecular Evolution (2006)
-
Initial sequence of the chimpanzee genome and comparison with the human genome
Nature (2005)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.