Abstract
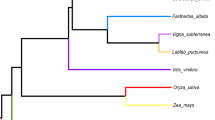
A central component of the endosymbiotic theory for the bacterial origin of the mitochondrion is that many of its genes were transferred to the nucleus. Most of this transfer occurred early in mitochondrial evolution1; functional transfer of mitochondrial genes has ceased in animals2. Although mitochondrial gene transfer continues to occur in plants3, no comprehensive study of the frequency and timing of transfers during plant evolution has been conducted. Here we report frequent loss (26 times) and transfer to the nucleus of the mitochondrial gene rps10 among 277 diverse angiosperms. Characterization of nuclear rps10 genes from 16 out of 26 loss lineages implies that many independent, RNA-mediated rps10 transfers occurred during recent angiosperm evolution; each of the genes may represent a separate functional gene transfer. Thus, rps10 has been transferred to the nucleus at a surprisingly high rate during angiosperm evolution. The structures of several nuclear rps10 genes reveal diverse mechanisms by which transferred genes become activated, including parasitism of pre-existing nuclear genes for mitochondrial or cytoplasmic proteins, and activation without gain of a mitochondrial targeting sequence.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Gray, M. W. The endosymbiont hypothesis revisited. Int. Rev. Cytol. 141, 233–357 (1992).
Boore, J. L. Animal mitochondrial genomes. Nucleic Acids Res. 27 , 1767–1780 (1999).
Adams, K. L. et al. Intracellular gene transfer in action: Dual transcription and multiple silencings of nuclear and mitochondrial cox2 genes in legumes. Proc. Natl Acad. Sci. USA 96, 13863 –13868 (1999).
Wolfe, K. H., Li, W. -H. & Sharp, P. M. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl Acad. Sci. USA 84, 9054–9058 (1987).
Laroche, J., Li, P., Maggia, L. & Bousquet, J. Molecular evolution of angiosperm mitochondrial exons and introns. Proc. Natl Acad. Sci. USA 94, 5722–5727 ( 1997).
Kadowaki, K., Kubo, N., Ozawa, K. & Hirai, A. Targeting presequence acquisition after mitochondrial gene transfer to the nucleus occurs by duplication of existing targeting signals. EMBO J. 15, 6652–6661 (1996).
Figueroa, P., Gómez, I., Holuigue, L., Araya, A. & Jordana, X. Transfer of rps14 from the mitochondrion to the nucleus in maize implied integration within a gene encoding the iron-sulphur subunit of succinate dehydrogenase and expression by alternative splicing. Plant J. 18, 601– 609 (1999).
Kubo, N., Harada, K., Hirai, A. & Kadowaki, K. A single nuclear transcript encoding mitochondrial RPS14 and SDHB of rice is processed by alternative splicing: Common use of the same mitochondrial targeting signal for different proteins. Proc. Natl Acad. Sci. USA 96, 9207–9211 (1999).
Wischmann, C. & Schuster, W. Transfer of rps10 from the mitochondrion to the nucleus in Arabidopsis thaliana: evidence for RNA-mediated transfer and exon shuffling at the integration site. FEBS Lett. 375, 152–156 (1995).
Kubo, N. et al. Transfer of the mitochondrial rps10 gene to the nucleus in rice: acquisition of the 5′ untranslated region followed by gene duplication. Mol. Gen. Genet. 263, 733– 739 (2000).
Nakagawa, T., Maeshima, M., Nakamura, K. & Asahi, T. Molecular cloning of a cDNA for the smallest nuclear-encoded subunit of sweet potato cytochrome c oxidase. Eur. J. Biochem. 191, 557–561 (1990).
Morikami, A., Ehara, G. & Yuuki, K. Molecular cloning and characterization of cDNAs for the γ-subunit and ε-subunit of mitochondrial F1F0 ATP synthase from sweet potato. J. Biol. Chem. 268, 17205–17210 (1993).
Braun, H.-P., Jansch, L., Kruft, V. & Schmitz, U. K. The ‘Hinge’ protein of cytochrome c reductase from potato lacks the acidic domain and has no cleavable presequence. FEBS Lett. 347, 90–94 (1994).
Long, M., de Souza, S. J., Rosenberg, C. & Gilbert, W. Exon shuffling and the origin of the mitochondrial targeting function in plant cytochrome c1 precursor. Proc. Natl Acad. Sci. USA 93, 7727–7731 (1996).
Herrmann, R. G. in Eukaryotism and Symbiosis (eds Schenk, H. E. A. et al.) 73–118 (Springer, Vienna, 1997).
Figueroa, P. et al. The gene for mitochondrial ribosomal protein S14 has been transferred to the nucleus in Arabidopsis thaliana. Mol. Gen. Genet. 262, 139–144 (1999).
Bensasson, D., Zhang, D.-X. & Hewitt, G. M. Frequent assimilation of mitochondrial DNA by grasshopper nuclear genomes. Mol. Biol. Evol. 17, 406 –415 (2000).
Palmer, J. D. The mitochondrion that time forgot. Science 275, 790–791 (1997).
Martin, W. et al. Gene transfer to the nucleus and the evolution of chloroplasts. Nature 393, 162–165 (1998).
Gray, M. W. Evolution of organellar genomes. Curr. Opin. Genet. Dev. 9, 678–687 (1999).
Swofford, D. L. PAUP*, Phylogenetic Analysis Using Parsimony (*and Other Methods) 4.0B3 edn (Sinauer Associates, Sunderland, MA, 2000).
Strimmer, K. & von Haeseler, A. Quartet puzzling: A quartet maximum likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13, 964–969 (1996).
Day, D. A., Neuburger, M. & Douce, R. Biochemical characterization of chlorophyll-free mitochondria from pea leaves. Aust. J. Plant Phys. 12, 219–228 (1985).
Neuberger, M., Journet, E., Bligny, R., Carde, J. & Douce, R. Purification of plant mitochondria by isopycnic centrifugation in density gradients of percoll. Arch. Biochem. Biophys. 217, 312–323 (1982).
Tanudji, M., Sjoling, S., Glaser, E. & Whelan, J. Signals required for the import and processing of the alternative oxidase into mitochondria. J. Biol. Chem. 274, 1286– 1293 (1999).
Knox, C., Sass, E., Neupert, W. & Pines, O. Import into mitochondria, folding, and retrograde movement of fumarase in yeast. J. Biol. Chem. 273, 25587–25593 ( 1998).
Soltis, P. S., Soltis, D. E. & Chase, M. W. Angiosperm phylogeny inferred from multiple genes as a tool for comparative biology. Nature 402, 402–404 (1999).
Acknowledgements
We thank W. Fischer, J. Logsdon, C. Parkinson, M. Rosenblueth, N. Schisler and K. Wolfe for reading the manuscript, and D. Swofford for allowing us to use a pre-release version of PAUP*. This study was supported by a United States Department of Agriculture graduate fellowship to K.L.A., grants to Y.L.Q. and J.D.P. from the NIH, and grants from the Australian Research Council to J.W.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Rights and permissions
About this article
Cite this article
Adams, K., Daley, D., Qiu, YL. et al. Repeated, recent and diverse transfers of a mitochondrial gene to the nucleus in flowering plants. Nature 408, 354–357 (2000). https://doi.org/10.1038/35042567
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35042567
This article is cited by
-
Horizontal gene transfer in eukaryotes: aligning theory with data
Nature Reviews Genetics (2024)
-
Both Conifer II and Gnetales are characterized by a high frequency of ancient mitochondrial gene transfer to the nuclear genome
BMC Biology (2021)
-
Foreign Plastid Sequences in Plant Mitochondria are Frequently Acquired Via Mitochondrion-to-Mitochondrion Horizontal Transfer
Scientific Reports (2017)
-
The transcriptome landscape of early maize meiosis
BMC Plant Biology (2014)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.