Abstract
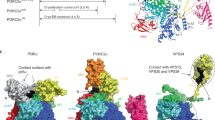
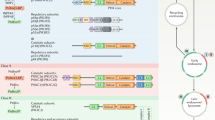
Phosphoinositide 3-kinases (PI3Ks) are ubiquitous lipid kinases that function both as signal transducers downstream of cell-surface receptors and in constitutive intracellular membrane and protein trafficking pathways. All PI3Ks are dual-specificity enzymes with a lipid kinase activity which phosphorylates phosphoinositides at the 3-hydroxyl, and a protein kinase activity. The products of PI3K-catalysed reactions, phosphatidylinositol 3,4,5-trisphosphate (PtdIns(3,4,5)P3), PtdIns(3,4)P2 and PtdIns(3)P, are second messengers in a variety of signal transduction pathways, including those essential to cell proliferation, adhesion, survival, cytoskeletal rearrangement and vesicle trafficking1,2. Here we report the 2.2 Å X-ray crystallographic structure of the catalytic subunit of PI3Kγ, the class I enzyme that is activated by heterotrimeric G-protein βγ subunits and Ras. PI3Kγ has a modular organization centred around a helical-domain spine, with C2 and catalytic domains positioned to interact with phospholipid membranes, and a Ras-binding domain placed against the catalytic domain where it could drive allosteric activation of the enzyme.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Toker,A. & Cantley,L. C. Signalling through the lipid products of phosphoinositide-3-OH kinase. Nature 387, 673–676 (1997).
Domin,J. & Waterfield,M. D. Using structure to define the function of phosphoinositide 3-kinase family members. FEBS Lett. 410, 91–95 (1997).
Stoyanov,B. et al. Cloning and characterisation of a G protein-activated human phosphoinositide-3 kinase. Science 269, 690–693 (1995).
Stephens,L. R. et al. The Gβγ sensitivity of a PI3K is dependent upon a tightly associated adaptor, p101. Cell 89, 105–114 (1997).
Krugmann,S., Hawkins,P. T., Pryer,N. & Braselmann,S. Characterizing the interactions between the two subunits of the p101/p110γ phosphoinositide 3-kinase and their role in the activation of this enzyme by Gβγ subunits. J. Biol. Chem. 274, 17152–17158 (1999).
Taylor,S. S. et al. Catalytic subunit of cyclic AMP-dependent protein kinase: structure and dynamics of the active site cleft. Pharmacol. Ther. 82, 133–141 (1999).
Dhand,R. et al. PI3-kinase is a dual specificity enzyme—autoregulation by an intrinsic protein serine kinase activity. EMBO J. 13, 522–533 (1994).
Stack,J. H. & Emr,S. D. Vps34p required for yeast vacuolar protein sorting is a multiple specificity kinase that exhibits both protein kinase and phosphatidylinositol-specific PI-3-kinase activities. J. Biol. Chem. 269, 31552–31562 (1994).
Wymann,M. P. et al. Wortmannin inactivates phosphoinositide 3-kinase by covalent modification of Lys-802, a residue involved in the phosphate transfer reaction. Mol. Cell. Biol. 16, 1722–1733 (1996).
Bondeva,T. et al. Bifurcation of lipid and protein kinase signals of PI3Kγ to the protein kinases PKB and MAPK. Science 282, 293–296 (1998).
Vanhaesebroeck,B. et al. Autophosphorylation of p110δ phosphoinositide 3-kinase: a new paradigm for the regulation of lipid kinases in vitro and in vivo. EMBO J. 18, 1292–1302 (1999).
Kim,K. & Cole,P. A. Measurement of a Brønsted nucleophile coefficient and insights into the transition state for a protein tyrosine kinase. J. Am. Chem. Soc. 119, 11096–11097 (1997).
Marshall,C. J. Ras effectors. Curr. Opin. Cell Biol. 8, 197–204 (1996).
Moodie,S. A. et al. Different structural requirements within the switch II region of the Ras protein for interactions with specific downstream targets. Oncogene 11, 447–454 (1995).
Rodriguez-Viciana,P. et al. Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell 89, 457–467 (1997).
Nassar,M. et al. the 2.2 Å crystal structure of the Ras-binding domain of the serine/threonine kinase c-Raf1 in complex with Rap1A and a GTP analogue. Nature 375, 554–560 (1995).
Huang,L., Hofer,F., Martin,G. S. & Kim,S.-H. Structural basis for the interaction of Ras with RalGDS. Nature Struct. Biol. 5, 422–426 (1998).
Essen,L.-O., Perisic,O., Lynch,D. E., Katan,M. & Williams,R. L. A ternary metal binding site in the C2 domain of phosphoinositide-specific phospholipase C-δ1. Biochemistry 36, 2753-2762 (1997).
Rao,V. D., Misra,S., Boronenkov,I. V., Anderson,R. A. & Hurley,J. H. Structure of type IIβ phosphatidylinositol phosphate kinase: a protein kinase fold flattened for interfacial phosphorylation. Cell 94, 829–839 (1998).
Groves,M. R., Hanlon,N., Turowski,P., Hemmings,B. A. & Barford,D. The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs. Cell 96, 99–110 (1999).
Vetter,I. R., Arndt,A., Kutay,U., Görlich,D. & Wittinghofer,A. Structural view of the Ran-importin β interaction at 2.3 Å resolution. Cell 97, 635–646 (1999).
Dennis,P. B., Fumagalli,S. & Thomas,G. Target of rapamycin (TOR): balancing the opposing forces of protein synthesis and degradation. Curr. Opin. Genet. Dev. 9, 49–54 (1999).
Leslie,A. G. W. Recent changes to the MOSFLM package for film and image plate data, in Joint CCP4 and ESF-EACMB Newsletter on Protein Crystallography Vol. 26 (Daresbury Laboratory, Warrington, UK, 1992).
CCP4. Collaborative Computing Project 4: A suite of programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994).
Terwilliger,T. C. & Berendzen,J. Automated structure solution for MIR and MAD. Acta Crystallogr. D 55, 849–861 (1999).
de La Fortelle,E. & Bricogne,G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 276, 472–494 (1997).
Jones,T. A., Zou,J.-Y., Cowan,S. W. & Kjeldgaard,M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Brünger,A. T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Nicholls,A., Sharp,K. A. & Honig,B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins Struct. Funct. Genet. 11, 281–296 (1991).
Yamaguchi,H. & Hendrickson,W. A. Structural basis for the activation of human lymphocyte kinase Lck tyrosine phosphorylation. Nature 384, 484–489 (1996).
Kraulis,P. J. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950 (1991).
Acknowledgements
We thank D. Lynch for technical assistance, S. Krugmann for PI3K assays, the staff of synchrotron beamlines ID2b, ID14-3 and ID14-4 at ESRF, IMCA-CAT at APS, X11 at EMBL Hamburg, Station 9.6 at Daresbury SRS, and Elettra, Italy, for help in synchrotron data collection. We thank P. Roversi and G. Bricogne for their assistance with BUSTER, R. Rubin and C. Humblet for access to the IMCA-CAT beamline, and P. Hawkins and A. Murzin for helpful discussions. C.R. was supported by the Deutsche Forschungsgemeinschaft. We are grateful for support from the BBSRC (via a senior fellowship to P. Hawkins), the British Heart Foundation, Parke-Davis and Onyx Pharmaceuticals (R.L.W.).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Walker, E., Perisic, O., Ried, C. et al. Structural insights into phosphoinositide 3-kinase catalysis and signalling. Nature 402, 313–320 (1999). https://doi.org/10.1038/46319
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/46319
This article is cited by
-
Oncogenic mutations of PIK3CA lead to increased membrane recruitment driven by reorientation of the ABD, p85 and C-terminus
Nature Communications (2023)
-
Development of selective inhibitors of phosphatidylinositol 3-kinase C2α
Nature Chemical Biology (2023)
-
Structural basis of phosphatidylinositol 3-kinase C2α function
Nature Structural & Molecular Biology (2022)
-
Role of PRKDC in cancer initiation, progression, and treatment
Cancer Cell International (2021)
-
The novel duplication HRAS c.186_206dup p.(Glu62_Arg68dup): clinical and functional aspects
European Journal of Human Genetics (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.