Abstract
Cellular stress responses can be activated following functional defects in organelles such as mitochondria and the endoplasmic reticulum. Mitochondrial dysfunction caused by loss of the serine protease HtrA2 leads to a progressive movement disorder in mice and has been linked to parkinsonian neurodegeneration in humans. Here, we demonstrate that loss of HtrA2 results in transcriptional upregulation of nuclear genes characteristic of the integrated stress response, including the transcription factor CHOP, selectively in the brain. We also show that loss of HtrA2 results in the accumulation of unfolded proteins in the mitochondria, defective mitochondrial respiration and enhanced production of reactive oxygen species that contribute to the induction of CHOP expression and to neuronal cell death. CHOP expression is also significantly increased in Parkinson's disease patients’ brain tissue. We therefore propose that this brain-specific transcriptional response to stress may be important in the advance of neurodegenerative diseases.
Similar content being viewed by others
Main
The serine protease HtrA2 (also known as Omi) was initially identified as a mammalian homolog of the Escherichia coli stress responsive proteases HtrA/DegP and DegS.1 Mammalian HtrA2 normally resides in the mitochondrial intermembrane space, with full-length HtrA2 being attached to the inner mitochondrial membrane. Initial studies have suggested that HtrA2 contributed to apoptosis in a manner analogous to pro-apoptotic Reaper family proteins present in Drosophila melanogaster.2 However, this view of HtrA2 function has been thrown into doubt by the phenotype of mice with loss of HtrA2 activity, which do not show decreased apoptosis, but die prematurely at about 30 days of age, displaying a progressive neurodegenerative disorder with parkinsonian features.3, 4
In humans, a polymorphism and a mutation in HtrA2 that affect its proteolytic activity have been found in Parkinson's disease (PD) patients.5 More recently, HtrA2 and the putative kinase PINK1 have been proposed to be components of a mitochondrial stress sensing pathway that might be defective in PD patients.6 HtrA2 may act to protect mitochondria from certain stresses, in a manner similar to the homologous stress-adaptive proteins DegP and DegS in bacteria,7, 8 and may be part of the mechanism underlying the well established but poorly understood link between mitochondrial dysfunction and neurodegenerative disease.9
Eukaryotic cells respond to diverse stress signals with programmes of nuclear gene expression designed to repair cellular damage or induce apoptosis. The endoplasmic reticulum (ER) specific or mitochondria-specific unfolded protein responses (UPRs) are activated when misfolded proteins accumulate in the respective organelle (UPRER10 and UPRmt11, 12). The integrated stress response (ISR) is a general stress response program that acts to modulate protein biosynthesis by integrating various types of stress stimuli. The UPRER itself causes activation of the ISR, whereas several other stresses such as oxidative stress, nutrient limitation and double-stranded RNA will activate this program without affecting the UPRER (Supplementary Figure S1). Activation of the ISR results in the induction of downstream genes, including the transcription factor CHOP (also known as GADD153 or DDIT3).13 Reactive oxygen species (ROS) inducing parkinsonian mimetic toxins have been shown to induce the activation of the UPRER with concomitant activation of the ISR.14, 15
On the basis of the hypothesis that loss of HtrA2 may contribute to enhanced mitochondrial stress, we have identified the stress-inducible transcription factor CHOP as being differentially regulated upon loss of this protease. CHOP is induced in HtrA2 knockout (KO) mice specifically in the brain, and epistasis analysis shows that its loss suppresses neuronal cell death in HtrA2 KO animals. Transcriptional profiling revealed a set of genes, including CHOP, that are specifically upregulated in HtrA2 KO mouse brains and that had previously been characterized to be induced in neuronal cells stimulated with neurotoxins,14, 15 transcripts related to activation of the ISR but not the UPRER. Additionally, analysis of CHOP and HERP transcripts in human brains showed a significant upregulation in a subset of patients suffering from idiopathic PD (IPD) further linking the concept of ISR activation to neurodegenerative processes. Loss of HtrA2 contributes to increased accumulation of unfolded components of the respiratory machinery in mitochondria, defects in the electron transport chain and to enhanced production of ROS. Antioxidant treatment of HtrA2 KO mice alleviates their phenotype and partially suppresses neurodegeneration, suggesting that an increase in ROS production upon loss of HtrA2 might contribute to the activation of a stress response of mitochondrial origin in the brain that promotes neurodegeneration.
Results
HtrA2 KO cells show increased sensitivity to mitochondrial stress characterized by enhanced CHOP expression
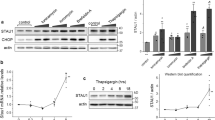
To test the contribution of HtrA2 to the transcriptional activation of a stress response, wild-type (WT) and HtrA2 KO primary mouse embryonic fibroblasts (MEFs) were challenged with an inducer of mitochondrial stress and their transcriptional response was monitored using microarray technology (experimental outline, Supplementary Figure S2a). WT and HtrA2 KO MEFs were chosen, as these cells have been shown earlier to display differential sensitivity to drug-induced stresses triggered by specific respiratory inhibitors such as rotenone, which specifically blocks complex I.4 This analysis revealed that in HtrA2 KO MEFs, the number of rotenone-induced transcriptional changes was significantly enhanced, suggesting that loss of HtrA2 results in an enhanced transcriptional response (Figure 1a). Among the genes most highly induced by rotenone treatment in HtrA2 KO cells was the transcription factor CHOP (Supplementary Table S1), which is known to be induced in response to various stresses such as activation of the UPRER, UPRmt and ISR.
Loss of HtrA2 results in a transcriptional stress response. (a) Rotenone induces a greater number of transcriptional changes in HtrA2 KO cells. Venn diagram representation of the overlap of probe sets regulated by rotenone in WT and HtrA2 KO MEFs. (b) CHOP is upregulated in immortalized HtrA2 KO MEFs by rotenone treatment. Short treatment with rotenone fails to induce CHOP expression in WT cells, whereas it results in approximately twofold induction in HtrA2 KO cells. Prolonged rotenone treatment leads to a more pronounced induction of CHOP in KO MEFs when compared with WT controls. MEFs were treated with vehicle, 25 μM rotenone for 3 h or 10 μM rotenone for 8 h. Transcript levels of CHOP are shown relative to vehicle treatment. Mean values±S.D.; n=3. Statistically significant values (two-tailed unpaired t-test) are indicated. (c) CHOP mRNA is induced by transient downregulation of HtrA2. HtrA2 was downregulated using siRNA in Neuro-2a neuroblastoma cells, and immortalized WT and HtrA2 KO MEFs. Transcript levels of HtrA2 and CHOP are shown relative to control siRNA. Mean values±S.D.; n=3. Statistically significant values (two-tailed unpaired t-test) relative to control siRNA are indicated. ** for P<0.01 and * for P<0.05
The increased induction of CHOP following rotenone treatment in HtrA2-deficient cells was confirmed by performing reverse transcriptase quantitative real-time PCR (qPCR) analysis on immortalized WT and HtrA2 KO MEFs (Figure 1b). Furthermore, downregulation of HtrA2 using siRNAs was sufficient to promote CHOP upregulation in the absence of any additional stress stimulus (Figure 1c; Supplementary Figure S3a). As a control, we treated HtrA2 KO MEFs with the HtrA2-specific siRNAs (Figure 1c). Taken together, the data indicated that the transcriptional response to rotenone-mediated stress was significantly enhanced in cells lacking HtrA2, suggesting that this could be due to a diminished threshold for mitochondrial stress in HtrA2 KO cells.
CHOP is specifically induced in the brains of HtrA2 KO mice
The observation that the neuroblastoma cell line Neuro-2a showed enhanced upregulation of CHOP when HtrA2 was silenced (Figure 1c) together with our earlier observations that HtrA2 KO mice show selective loss of neuronal cells4 led us to investigate the transcriptional profile of brain tissue lacking HtrA2. Neurons in HtrA2 KO mice might be subjected to enhanced levels of mitochondrial stress even in the absence of exogenous stimuli. We therefore proceeded to compare transcript levels in brain tissue from the cortex of WT and HtrA2 KO mice (Supplementary Figure S2b). We chose a cortical region as there is no evidence of neuronal loss in this region enabling us to compare identical cellular populations. Transcriptional profiling revealed a significant number of genes with altered expression in the cortex of HtrA2 KO mice (Figure 2a; Supplementary Table S2).
CHOP expression is specifically induced in brains of HtrA2 KO mice. (a) Transcriptional profiling of mitochondrial stress signaling in HtrA2 KO mice. Volcano plot representation of all probe sets. Probe sets passing the 0.05 FDR threshold are highlighted in dark grey; arrows indicate probe set for CHOP 1 and HtrA2.2 X axis: KO/WT log2 ratio for each probe set, dotted vertical lines show twofold up- or downregulation of expression. Y axis: FDR in −log(10) units, horizontal lines show 0.01, 0.05 and 0.1 FDR threshold. (b) CHOP is induced in different brain regions of HtrA2 KO mice. WT and HtrA2 KO mouse brains were dissected into basal ganglia (BG), cortex (CX), cerebellum (CB) and substantia nigra (SN) at P28. Transcript levels of CHOP are shown relative to WT. (c) CHOP expression is specifically upregulated in HtrA2 KO brain tissue. Brain, heart, kidney, liver and skeletal muscle tissue were isolated from WT and HtrA2 KO mice at P22. Transcript levels of CHOP are shown relative to WT. (d) Time course of CHOP upregulation in different brain regions of HtrA2 KO mice. Transcript levels of CHOP in HtrA2 KO brain tissue are shown relative to WT at the respective postnatal day. All data are presented as mean values±S.D.; n=2–3 in each group. Statistically significant values (one-way (b, c) or two-way (d) ANOVA with Bonferroni's post-test) relative to WT are indicated. *** for P<0.001, ** for P<0.01 and * for P<0.05
Interestingly, the transcription factor CHOP was among the most highly upregulated transcripts in cortex tissue from HtrA2 KO mice. Extending this analysis, qPCR measurements revealed that CHOP expression was significantly upregulated in HtrA2 KO mice in all brain regions tested (Figure 2b). However, CHOP expression was not upregulated in any other tissue of HtrA2 KO mice tested (Figure 2c), suggesting that the brain displayed a selective vulnerability to loss of HtrA2 function. Analysis of animals heterozygous for HtrA2 revealed that CHOP expression was comparable to that of WT animals (data not shown).
To determine whether CHOP upregulation preceded the movement disorder of HtrA2 KO mice, the levels of CHOP mRNA were monitored in specific brain regions isolated before (postnatal days P10–17) and after (P28) development of the phenotype. CHOP was selectively upregulated first in the basal ganglia at P15. This upregulation extended to the cortex at P17 and finally the cerebellum at P28 (Figure 2d). The increase of CHOP expression in the brains of HtrA2 KO mice appears progressive, with the earliest induction coinciding with the area where neurodegeneration is detected. Interestingly, the brain areas affected during this progressive CHOP upregulation are similar to those described for staging the brain pathology of sporadic PD patients.16
CHOP contributes to striatal neurodegeneration in HtrA2 KO mice
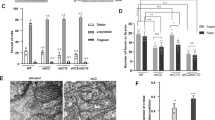
To determine the contribution of CHOP to the neuronal cell loss observed in HtrA2 KO mice, we generated mice deleted for both genes. Heterozygotes for both HtrA2 and CHOP mutant alleles were intercrossed and F2 progeny born with the expected Mendelian inheritance, indicating that deletion of both CHOP and HtrA2 does not result in embryonic lethality (data not shown). Mice lacking both CHOP and HtrA2 failed to gain weight (Figure 3a), suggesting that CHOP deletion does not affect the pleotropic effects caused by deletion of HtrA2. HtrA2 KO mice are characterized by loss of striatal neurons; however, no cellular loss has been detected in the substantia nigra (Martins et al.4 and Supplementary Figure S4a). Levels of tyrosine hydroxylase are similar in WT and HtrA2 KO mice (Supplementary Figure S4b) and levels of striatal dopamine (DA) and its metabolite dihydroxyphenylacetic acid are not significantly altered in HtrA2 KO mice (Supplementary Figure S4c and S4d). Given the brain specificity of CHOP upregulation, we compared the neuronal cell loss in HtrA2 KO and HtrA2 KO/CHOP KO littermates. Loss of CHOP in HtrA2 KO background results in a decrease in striatal neurodegeneration (Figure 3b), although increased GFAP staining of astrocytes suggests that astrogliosis is occurring and neurodegeneration has been initiated. Quantitation of the density of NeuN-positive neurons and apoptotic cells (active caspase-3-positive cells) showed that the selective loss of striatal neurons and enhanced apoptosis observed in HtrA2 KO mice is partially suppressed in the absence of CHOP (Figure 3c and d).
CHOP contributes to neuronal cell death mediated by loss of HtrA2. (a) HtrA2 KO and HtrA2 KO/CHOP KO mice show similar growth curves. Average body weights of indicated mice. Mean values±S.E.M.; n=3. (b) Features of localized striatal degeneration revealed by NeuN immunohistochemistry at P30 in control (HtrA2 WT and HET), HtrA2 KO and double KO mice. Upper panels, NeuN staining; lower panels, GFAP staining (green) of astrocytes and Hoechst staining (purple) of cell nuclei. Scale bar corresponds to 400 μm. (c) Average number of NeuN-positive neurons in the area affected by HtrA2 loss at P30 in control, HtrA2 KO and double KO mice. Statistically significant values (one-way ANOVA with Bonferroni's multiple comparison test) relative to WT are indicated. (d) Average number of active caspase-3-positive cells in the area affected by HtrA2 loss at P30 in control, HtrA2 KO and double KO mice. Two-tailed t-test assuming unequal variance relative to HtrA2 KO are indicated. *** for P<0.001 and * for P<0.05
Activation of the ISR following loss of HtrA2
To identify genes that might be part of a stress response to loss of HtrA2, we compared expression data from MEFs after rotenone treatment with that obtained from WT and HtrA2 KO cortex samples. We tested the significance of this comparison using gene set enrichment analysis (GSEA).17 When rotenone-regulated genes in MEFs (gene set: MEF rotenone) were tested against gene expression in HtrA2 KO brain, the GSEA indicated a robust statistical significance (Table 1), revealing an overlapping transcriptional stress response both in vitro and in vivo (Table 2).
It has been reported that cultured neuronal cells treated with parkinsonian mimetics activate a transcriptional program characteristic of activation of the UPRER and ISR, including the transcription factor CHOP.14, 15 Furthermore, CHOP is upregulated in neurotoxin models of parkinsonism in vivo.19 We noted that genes induced in neuronal cells by both 6-OHDA (6-hydroxydopamine) and MPP+ (1-methyl-4-phenylpyridinium) were characteristic for activation of the ISR (gene set: neurotoxin treatment (ISR)), whereas only 6-OHDA would additionally activate genes characteristic for the UPRER (e.g. Grp58, Xbp1) (gene set: neurotoxin treatment (UPRER)).14 We thus examined whether the transcriptional changes in the brains of HtrA2 KO mice were comparable with the data sets derived from in vitro neurotoxin models. GSEA revealed high statistical significance between genes characteristic for the ISR, but not the UPRER, which are upregulated by these neurotoxins in vitro and genes upregulated in brains of HtrA2 KO mice (Tables 1 and 2), suggesting that loss of HtrA2 in vivo leads to activation of the ISR, but not the UPRER in the brain.
To further confirm the presence of ISR-characteristic genes upregulated in HtrA2 KO brain, we compared our data set with data from a study where WT and ATF4-deficient MEFs were treated with tunicamycin.18 In WT cells, tunicamycin treatment results in activation of the UPRER and concomitantly the ISR; however, in ATF4-deficient cells only the UPRER is activated and ATF4-dependent ISR transcripts are suppressed. GSEA showed a robust statistical significance (Table 1; gene set: ATF4 target genes), confirming that transcripts related to activation of the ISR are upregulated in brain tissue from HtrA2 KO mice (Table 2).
Previously, CHOP upregulation has been implicated in a mitochondria-specific stress response (UPRmt) leading to increased expression of mitochondrial chaperones, including Hsp60, Hsp10 and ClpP.12 Although none of these mitochondrial proteins was differentially expressed in our GeneChip analysis, we measured their expression by qPCR. The transcripts of ATF3 and HERP, both proteins that can be induced by the ISR, were found to be upregulated in brains from HtrA2 KO mice, but neither Hsp60 nor ClpP transcripts were differentially expressed (Figure 4a), suggesting that CHOP induction in the brain fails to induce any mitochondria protective genes. Similarly to CHOP, ATF3 and HERP expression was upregulated in the brains of HtrA2 KO mice, but not in other tissues tested (Figure 4b).
Activation of the ISR in brains from HtrA2 KO mice and IPD patients. (a) ATF3 and HERP are upregulated in HtrA2 KO brain tissue, but not Hsp60 or ClpP. Basal ganglia were dissected from brains of WT and HtrA2 KO mice at P27. Transcript levels are shown relative to WT. Mean values±S.E.M.; n=3–5 in each group. (b) ATF3 and HERP expression are specifically upregulated in HtrA2 KO brain tissue. Brain, heart, kidney, liver and skeletal muscle tissue were isolated from WT and HtrA2 KO mice at P22. Transcript levels of ATF3 and HERP are shown relative to WT. Mean values±S.D.; n=3 in each group. Statistically significant values (one-way ANOVA with Bonferroni's post-test) relative to WT are indicated. (c) CHOP and HERP are significantly upregulated in a subset of IPD patients. Transcript levels are shown relative to one control patient. Means are indicated by the horizontal bars. Purple dots correspond to a selected set of IPD patients displaying levels of the CHOP transcript above the mean value. P-values (two-tailed t-test assuming unequal variance) relative to control are indicated. *** for P<0.001, ** for P<0.01 and * for P<0.05
The data reported so far suggest that HtrA2 deficiency contributes to activation of the ISR specifically in brain tissue of these mice. To investigate whether such a transcriptional response might be present in humans suffering from neurodegenerative disorders, we analyzed brains from PD patients as HtrA2 has been implicated in this disease.5, 6 Using qPCR, we compared the levels of CHOP and HERP mRNA expression in a group of control brains and a group of brains that fulfilled agreed criteria for the diagnosis of IPD. This revealed a relative increase in the ISR genes CHOP and HERP in a number of IPD patient brains compared with the control group (Figure 4c). Measuring the expression levels of the normalizer transcripts RPS18 and SDHA revealed no significant difference between control and IPD patient groups (Figure 4c), showing that this increased expression is specific to the ISR transcripts measured in this analysis.
Loss of HtrA2 KO results in the accumulation of unfolded proteins in mitochondria and defective respiration
We investigated whether loss of HtrA2 led to protein misfolding in the mitochondria by analyzing detergent-insoluble mitochondrial proteins.12 The enrichment of several proteins in HtrA2 KO mitochondria compared with WT (Figure 5a), indicated that these proteins are present as misfolded species in HtrA2 KO mitochondria. Given the close proximity of HtrA2 to the mitochondrial inner membrane,20 we determined whether loss of HtrA2 affects the correct folding of abundant inner membrane proteins. Interestingly, this revealed a significant increase in detergent-insoluble subunits of respiratory chain complexes in HtrA2 KO mitochondria (Figure 5b) under gentle lysis conditions.21 Additionally, given that the bacterial homolog of HtrA2 is involved in the correct folding of outer membrane β-barrel porins, we detected the presence of slower migrating forms of VDAC in HtrA2 KO brain mitochondria (Figure 5b, asterisk), consistent with enhanced misfolding of this porin. We failed to see incorrect folding of other mitochondrial proteins including SOD2 (Figure 5b) and DJ-1 (data not shown).
Defective mitochondria in HtrA2 KO mice and IPD patients. (a) Detection of misfolded proteins in mitochondria derived from HtrA2 KO mice. Mitochondria were lysed in the indicated concentrations of NP-40 or in 2% SDS under gentle lysis conditions as described,21 insoluble proteins were isolated, separated by SDS-PAGE and visualized by silver staining. Arrowheads indicate NP-40-insoluble proteins enriched in mitochondria lacking HtrA2. (b) Detection of protein misfolding in mitochondria derived from HtrA2 KO mice. Mitochondria were processed as before and proteins were detected by western blot using the indicated antibodies. Asterisk indicates the position of slower migrating VDAC. (c) Mitochondria isolated from brains of WT and HtrA2 KO mice were assayed for complex I- and II-dependent respiration. (d) Mitochondria isolated from brains of WT and HtrA2 KO mice were assayed for complex IV-dependent respiration either in the absence or presence of exogenous cytochrome c. (e) Frozen cortex brain tissue prepared from WT and HtrA2 KO mice was used to measure uncoupled respiration. All data are represented as mean values±S.D.; n>4 in each group. Statistically significant values (two-tailed unpaired t-test) are indicated. (f) Similar amounts of mtDNA are found in mitochondria from brains of WT and HtrA2 KO mice. Mean values±S.D.; n=3 in each group. (g) Levels of respiratory complexes in post-mortem brain lysates from control and IPD patients. Respiratory complexes were detected by western blot analysis using the indicated antibodies. Equivalent sample loading was confirmed by detection of GAPDH. *** for P<0.001, ** for P<0.01 and * for P<0.05
To estimate the effect of misfolded respiratory complexes in HtrA2 KO cells, we measured their activity by high-resolution respirometry, using isolated brain mitochondria and cortex tissue samples from WT and HtrA2 KO mice at P30. Coupled mitochondria derived from HtrA2 KO mice showed a significant reduction in oxygen consumption. A decrease in respiration of 40–50% was observed using substrates of complexes I and II (Figure 5c), as well as substrates of complex IV (Figure 5d). A generalized respiratory dysfunction was thus present in HtrA2 KO brain mitochondria, consistent with the presence of unfolded subunits of complexes I–IV (Figure 5b). This dysfunction was confirmed by measuring respiratory activities under uncoupled conditions with whole cortical brain (Figure 5e). Addition of cytochrome c with complex IV substrates did not restore the respiratory activity in HtrA2 KO mitochondria, thereby excluding cytochrome c availability as a potential limiting factor in the decreased respiratory activity (Figure 5d and e).
The mtDNA content of WT and HtrA2 KO brain analyzed by qPCR failed to reveal significant differences (Figure 5f), suggesting that the deficit in respiration in HtrA2 KO does not reflect a general loss of mtDNA.
To address whether induction of ISR components CHOP and HERP were linked to any mitochondrial defect in the IPD patient cohort (Figure 4c), we selected a subset of IPD patients displaying significantly higher levels of these transcripts and assessed the levels of respiratory complexes in brain samples. This revealed a significant decrease in subunits of respiratory complexes in cortex from several IPD patients when compared with controls (Figure 5g).
Loss of HtrA2 contributes to increased levels of ROS
We hypothesized that defective electron transport in HtrA2 KO mitochondria leads to increased oxidative stress, explaining the induction of the ISR. Therefore, we measured the intracellular concentration of ROS in immortalized WT and HtrA2 KO MEFs treated with the neurotoxin 6-OHDA, showing a marked increase in H2DCF-DA (2′–7′-dichlorofluorescein diacetate)-positive cells derived from HtrA2 KO animals when compared with WT by flow cytometry (Figure 6a). HtrA2 KO cells treated with 6-OHDA displayed increased levels of superoxide, measured using MitoSOX, when compared with WT cells (Figure 6b), confirming the mitochondrial origin of the ROS. Consistently, we observed increased accumulation of ROS and mitochondrial superoxide in immortalized HtrA2 KO MEFs following other treatments known to induce oxidative stress and/or related to neurotoxicity, such as H2O2 and rotenone (data not shown). Next, we re-introduced HtrA2 using a lentiviral expression system and confirmed this by western blot (Supplementary Figure S3b). Re-expression of HtrA2 in HtrA2 KO MEFs suppressed ROS generation following 6-OHDA treatment (Figure 6c). To extend our analysis to primary cells, we compared levels of ROS in primary MEFs (Figure 6d) and T cells from control and HtrA2 KO mice (Figure 6e). This confirmed that loss of HtrA2 results in enhanced production of oxygen radicals in cells treated with known inducers of cellular ROS.
Loss of HtrA2 leads to increased accumulation of ROS. (a) Accumulation of ROS is enhanced in HtrA2 KO cells. Immortalized WT and HtrA2 KO MEFs were stained with H2DCF-DA to measure intracellular ROS, treated with increasing concentrations of 6-OHDA for 3 h and analyzed by flow cytometry. (b) Increased accumulation of superoxide in HtrA2 KO mitochondria. Immortalized WT and HtrA2 KO MEFs were loaded with the mitochondrial superoxide indicator MitoSOX, treated with increasing concentrations of 6-OHDA for 3 h and analyzed by fluorescence microscopy (Cellomics KSR reader). (c) Restoration of HtrA2 expression reduces the accumulation of ROS as compared with non-restored cells. Immortalized HtrA2 KO MEFs and MEFs where HtrA2 expression was restored were stained with H2DCF-DA. (d) Primary WT, HET and HtrA2 KO MEFs were stained with H2DCF-DA and treated for 3 h (6-OHDA 50 μM, rotenone 10 μM and TNFα 25 ng/ml). (e) Thymocytes isolated from WT and HtrA2 KO animals were stained with H2DCF-DA and treated with increasing concentrations of 6-OHDA or rotenone for 3 h. (f) Immortalized WT and HtrA2 KO MEFs were pretreated with vehicle, NAC (100 μM, 2.5 h) and treated with 6-OHDA (50 μM, 16 h) or both, stained with propidium iodide (PI) to measure cell death and analyzed by flow cytometry. All data are presented as mean values±S.D.; n=3 in each group. Statistically significant values (one-way (a, b, c, f) or two-way (d, e) ANOVA with Bonferroni's post-test) relative to WT are indicated. *** for P<0.001, ** for P<0.01 and * for P<0.05
Cells derived from mice where the proteolytic activity of HtrA2 has been abolished display enhanced sensitivity to stress stimuli.3, 4 To determine the relevance of ROS accumulation for cell death sensitivity of HtrA2 KO cells, we investigated the effect of antioxidants on the viability of WT and HtrA2 KO MEFs (Figure 6f). This revealed a marked suppression of cell death (PI-positive cells) induced by 6-OHDA in HtrA2 KO cells pretreated with the antioxidant N-acetyl-L-cysteine (NAC). To determine whether the increased production of ROS in HtrA2 KO cells is related to a deficit in proteins involved in the response to oxidative stress, we performed western blot analysis for the antioxidant enzymes SOD1 and SOD2, revealing comparable levels in WT and HtrA2 KO (Supplementary Figure S3c). We compared the oxidative status of DJ-1, a sensor of oxidative stress, in 2D gels of mitochondrial lysates from WT and HtrA2 KO mice, which failed to detect significant alteration in this protein (Supplementary Figure S3d). Additionally, qPCR analysis of the mRNA levels of antioxidant enzymes GSTA4, SOD1 and SOD2 did not detect significant alterations in HtrA2 KO tissues (data not shown).
Finally, we explored whether loss of HtrA2 resulted in increased oxidative damage at the membrane level in vivo. To this end, we performed mass spectrometric analysis of mitochondrial lipids from murine liver and brain.22 Detailed analysis of electrospray profiles revealed an increase in peroxidized lipid species in both brain and liver mitochondria from HtrA2 KO mice (Table 3), thereby suggesting an enhanced ROS-mediated damage to membrane lipids in these animals. Consequently, loss of HtrA2 appears to trigger accumulation of oxidative stress at both the protein and lipid levels.
Antioxidants suppress striatal neurodegeneration in HtrA2 KO mice
We investigated whether increased ROS were causally implicated in the severe phenotypic defects in HtrA2 KO by treating the animals daily with the antioxidants NAC and TEMPOL. We did not detect significant difference in weight loss in HtrA2 KO mice treated with antioxidants when compared with the controls (Figure 7a), suggesting that in vivo, antioxidant treatment fails to suppress the pleotropic effects caused by loss of HtrA2. However, antioxidant treated HtrA2 KO mice displayed a significant decrease in the akinetic phenotype as assessed by open-field locomotor assays (Figure 7b). Next, we analyzed their striatal neurodegenerative phenotype, revealing that treatment of HtrA2 KO animals with NAC resulted in a reduction of the loss of NeuN-positive neurons and in the number of apoptotic cells (Figure 7c–e). GFAP immunostain revealed that even though antioxidant treatment suppressed the loss of NeuN-positive neurons, it failed to abolish glial cell invasion (Figure 7c).
Antioxidants suppress neurodegeneration in HtrA2 KO mice. (a) Antioxidant treatment fails to suppress failure to gain weight in HtrA2 KO mice. Average body weight of control (HtrA2 WT and HET) and HtrA2 KO animals fed with either milk or milk supplemented with NAC. Mean values±S.E.M.; n=3. (b) Effect of antioxidants on the locomotor activity of male HtrA2 KO mice measured at P29. Data represented as scatter plot. (c) Features of localized striatal degeneration at P30 in control, HtrA2 KO mice and HtrA2 KO mice treated with NAC. Upper panels, NeuN staining, lower panels, immunofluorescence images showing GFAP staining (green) of astrocytes and Hoechst staining (purple) of cell nuclei. Scale bar corresponds to 400 μm. (d) NAC treatment of HtrA2 KO mice rescues neuronal loss. Density of NeuN-positive neurons in the area affected by HtrA2 loss at P30 in control, HtrA2 KO and HtrA2 KO mice treated with NAC. (e) NAC treatment of HtrA2 KO mice suppresses caspase-3 activation. Density of cells staining positive for cleaved caspase-3 in control, HtrA2 KO and HtrA2 KO mice treated with NAC. (f) Immortalized WT and HtrA2 KO MEFs were treated with vehicle, NAC (10 mM), 6-OHDA (25 μM) or a combination of both for 18 h. Transcript levels of CHOP are shown relative to vehicle control for same genotype. Data in (d, e, f) are represented as mean values±S.D.; n=3 in each group. Statistically significant values (one-way ANOVA with Bonferroni's post-test) relative to vehicle-treated WT are indicated for (b, d, e, f). *** for P<0.001, ** for P<0.01 and * for P<0.05
We next tested the effects of antioxidants on the stress-induced transcriptional upregulation of CHOP. In cultured MEFs, pretreatment of cells with NAC led to the suppression of CHOP expression induced by 6-OHDA (Figure 7f), further confirming earlier studies23, 24 showing a role for mitochondrial ROS in the transcriptional upregulation of CHOP. Given that our data suggest that CHOP plays a significant role in neuronal cell death following induction of ROS, we next compared cell death rates in primary neurons derived from WT, HtrA2 KO, CHOP KO or HtrA2 KO/CHOP KO mice challenged with 6-OHDA in the presence or absence of antioxidants (Supplementary Figure S5). This revealed that deletion of CHOP is sufficient to protect neurons from ROS inducers even in the absence of antioxidants.
Discussion
In the present study, we have determined that loss of murine HtrA2 leads to compromised mitochondrial respiration and damage with accumulation of peroxidized lipids and unfolded proteins in this organelle. Such damage initiates a transcriptional stress response including the activation of ISR genes, which in vivo is specific to the brain. The bacterial protease DegS is a critical mediator of a transcriptional stress response initiated in the periplasmic space that mediates a transcriptional program in response to unfolded periplasmic proteins.25 In mammalian mitochondria, the absence of HtrA2 might compromise such a stress response inherited from bacteria and lead to enhanced mitochondria-initiated stress, possibly due to poor protein quality control in this organelle. Such failure in quality control, particularly at the level of the mitochondrial inner membrane, might affect normal electron transport through the respiratory chain. Consistent with such a model, our data suggest that absence of HtrA2 results in a generalized respiratory dysfunction.
It is attractive to propose that lack of HtrA2 might, directly or indirectly, have a deleterious effect on the proper folding of a number of mitochondrial proteins, in particular components or modulators of respiratory complexes at the mitochondrial membrane. This could possibly reflect a failure to clear unfolded respiratory complex proteins when the protease activity of HtrA2 is absent. The generalized damage to mitochondrial respiration resulting from loss of HtrA2 leads to excessive production of ROS and consequent accumulation of oxidative damage. Importantly, we report here that this damage occurs also at the level of mitochondrial membrane lipids, similarly to what has been documented in PD patients.26
Transcriptional consequences of enhanced mitochondrial stress triggered by loss of HtrA2
The induction of CHOP expression has been observed earlier in vitro when unfolded proteins accumulate in the mitochondria:12 activation of the UPRmt leads to a CHOP-dependent upregulation of mitochondrial chaperones including Hsp60, Hsp10 and ClpP, forming a feedback mechanism that acts to clear mitochondria of unfolded proteins. Our study provides the first in vivo evidence pointing toward an important role for CHOP in a response initiated through mitochondrial stress. However, we failed to detect any changes in Hsp60 or ClpP, suggesting that the transcriptional changes in the brain of HtrA2 KO mice are temporally or qualitatively distinct from activation of the reported UPRmt.12
The transcription factor CHOP has been implicated as a mediator of apoptosis in the context of oxidative stress,27, 28 and was shown to be a target of mitochondrial ROS in cultured cells.23 Recently, oxidative stress elicited through 6-OHDA treatment was shown to lead to the induction of CHOP,24 therefore placing the generation of ROS upstream of transcriptional changes. However, it is interesting to note that we detected an increase in peroxidized lipids in mitochondria obtained from both livers and brains of HtrA2 KO mice, whereas the upregulation of CHOP was selective to the brain and not observed in the liver or any other organ. We therefore reason that any ROS-dependent induction of CHOP might be selective to particular cell lineages.
We determined that a transient stimulus with neurotoxins such as rotenone, 6-OHDA or MPP+ modulates a similar subset of genes found upregulated in the brain of HtrA2 KO mice, namely genes of the ISR. However, target genes of the UPRER were not significantly modulated. In agreement with this, CHOP upregulation was detected in animals treated with neurotoxins, but no evidence for UPRER activation could be detected,19 suggesting that also these drug-induced animal models of parkinsonism might primarily be characterized by activation of the ISR alone. It is noted that oxidative stress has been shown earlier to lead to induction of the ISR and concomitant upregulation of CHOP expression without activation of the UPRER in neuronal cells,28 suggesting that deregulation of this pathway might be common in the development of neurodegenerative diseases.
In response to ER stress, IRE1, ATF6 and PERK are activated, with PERK leading to induction of the ISR, which can also be activated by other stresses, such as oxidative stress, starvation, viral infection and so on (Supplementary Figure S1). For ER stress, it has recently been shown that IRE1 signaling is protective but transient, whereas persistent activation of PERK and the ISR in the absence of sustained IRE1 signaling leads to cell death in a model of retinitis pigmentosa caused by mutant rhodopsin.29 In HtrA2 KO mice, we similarly observe ISR activation/CHOP induction leading to enhanced neuronal cell loss, although we failed to detect any evidence of upregulation of the ER chaperones such as BiP (data not shown).
CHOP as a molecular marker and effector of parkinsonian neurodegeneration
It is striking that the transcriptional activation of CHOP in HtrA2 KO mice was detected exclusively in the brain even though loss of HtrA2 leads to generalized cellular dysfunction.4 This could potentially mirror a higher level of stress in brain due to an increased demand for mitochondrial function or a decreased capacity to cope with the stress elicited by loss of HtrA2 function. CHOP is upregulated in neuronal tissues of HtrA2 KO mice, even in areas where no neuronal degeneration is observed, suggesting that CHOP might represent an early marker of mitochondrial dysfunction that precedes any cellular loss. Significantly, we obtained evidence for enhanced expression of ISR genes CHOP and HERP in a subset of IPD patients. Additionally, we detected decreased levels of respiratory complexes in this patient subset, suggesting that mitochondria dysfunction or possibly mitochondrial loss might be linked to the induction of these genes. Given that CHOP upregulation is likely to precede neurodegeneration, CHOP may be an early indicator of parkinsonian neurodegeneration in humans.
Deletion of CHOP delays the neurodegenerative process of HtrA2 KO mice, in line with recent studies showing that CHOP plays an important role in neurodegeneration triggered by ischemia30 as well as in a mouse model for Charcot–Marie–Tooth 1B neuropathy.31 It has recently been demonstrated that HtrA2 is part of the mitochondrial complex containing the protease Parl and the Bcl-2-family-related protein Hax-1. This complex has been proposed to regulate the levels of Bax and therefore susceptibility to apoptotic cell death. This study showed that deficiencies in Bax, Bak or Bim suppressed enhanced cell death of HtrA2-deficient lymphocytes but not the neurodegenerative phenotype caused by loss of HtrA2,32 indicating that the mechanisms of neuronal cell death in the absence of HtrA2 are distinct from those present in other cells or tissues. In the present study, we provide compelling data supporting this divergence in cell death mechanisms, demonstrating that cell death in the brains of HtrA2 KO animals involves transcriptional changes in gene expression and CHOP activation.
In the absence of HtrA2, cumulative build-up of oxidative damage occurs. Failure to control the effects of oxidative stress causes further amplification of ROS accumulation by impairing normal respiratory processes. Consequently, activation of stress response-specific genes characteristic for an ISR takes place presumably to protect against stress. However, activation of antioxidant mechanisms or induction of protective mitochondrial chaperones could not be detected. Thus, prolonged activation of stress transcriptional factors in the absence of upregulation of protective mechanisms appears to have a detrimental effect and contributes to cell death.
The HtrA2 KO mouse is the first genetic mouse model suffering from a neurodegenerative phenotype that shows upregulation of CHOP, interestingly specifically in the brain. A model can thus be proposed whereby HtrA2 modulates the levels of unfolded proteins in the mitochondria and oxidative stress. Loss of HtrA2 compromises this pathway leading to enhanced accumulation of unfolded proteins, increased oxidative stress, resulting in expression of CHOP and modulation of a number of other genes leading to neuronal cell death (Figure 8). This mouse model of parkinsonism has permitted insights into human PD, including our observation of upregulation of CHOP expression in brain tissue of IPD patients. Our earlier studies indicate that the putative kinase PINK1 acts upstream of HtrA2 in a mitochondrial stress sensing pathway that might be defective in PD, and the possibility exists that the products of other genes implicated in PD might also influence this pathway. It is attractive to propose that activation of ISR genes, including CHOP might be a common feature in PD, in particular when mitochondrial dysfunction is the main causative agent. We suggest that this transcriptional response to stress may play an important role in the progression of neurodegenerative diseases and could present new therapeutic opportunities.
Model for HtrA2 function. To the left, the DegS-dependent periplasmic stress response pathway of Gram-negative bacteria is outlined.8 To the right, a model for mammalian HtrA2, whereby HtrA2 function is necessary to prevent the accumulation of unfolded proteins and oxidative stress. Therefore, loss of HtrA2 leads to an increase in unfolded proteins and oxidative stress, resulting in activation of the ISR characterized by upregulation of expression of CHOP and a number of other genes, contributing to neuronal cell death. Antioxidants suppress generation of oxidative stress and alleviate the loss of neuronal cell in HtrA2 KO mice. Part of the model has been suggested by the present study (gray hexagon). HtrA2 and the putative kinase PINK1 have been proposed to be components of a mitochondrial stress sensing pathway that might be defective in PD patients6
Materials and Methods
Isolation of mitochondria
Mitochondria were purified from fresh mouse brains and livers obtained from WT and HtrA2 KO mice in homogenization buffer (10 mM HEPES pH 7.4, 50 mM sucrose, 0.4 M mannitol, 10 mM KCl and 1 mM EGTA) using a two-step differential centrifugation at 600 × g and 3000 × g, respectively, for 15 min at 4 °C. The mitochondria-enriched fraction was resuspended in a buffer containing 20 mM HEPES pH 7.4, 0.4 M mannitol, 10 mM NaH2PO4, 0.5 mM EGTA, and used for respirometry experiments or snap frozen in liquid nitrogen and preserved at −80 °C for further analysis.
Mitochondrial fractionation
Frozen mitochondria isolated from WT and HtrA2 KO mice were resuspended at a final concentration of 1 mg/ml in lysis buffer (25 mM Tris-HCl pH 7.4, 300 mM NaCl, 5 mM EDTA, 1 μg/ml chymostatin, 1 μg/ml leupeptin, 1 μg/ml pepstatin A and 1 mM PMSF). Subsequently, an equal volume of lysis buffer containing either SDS (4%) or NP-40 (2, 1, 0.4 or 0.2%) was added to each sample. Samples were further incubated on ice for 10 min. The NP-40-insoluble aggregates were pelleted by centrifugation (20 000 × g) at 4 °C and resuspended in NuPAGE LDS Sample Buffer (Invitrogen, Paisley, UK). Throughout the protein extractions, gentle lysis conditions were used21 (samples were kept on ice in the absence of reducing agents). Proteins were resolved in NuPAGE 4–12% gradient gels (Invitrogen) and visualized by silver staining (Invitrogen) or separated on 12% SDS-PAGE gels for western blot analysis using the indicated antibodies.
Flow cytometry
MEFs and primary thymocytes were analyzed using a FACS Calibur Instrument (Becton Dickinson, Mountain View, CA, USA). Cell viability was assessed by propidium iodide staining. ROS levels were measured by staining the cells with 50 μM H2DCF-DA (Molecular Probes, Paisley, UK) for 45 min at 37 °C in HEPES buffer (25 mM HEPES, 15 mM glucose, 120 mM NaCl, 5 mM KCl, 1.8 mM CaCl2, 2 mM MgCl2, pH 7.4) followed by treatment with 6-OHDA (25, 50 and 100 μM), rotenone (1–40 μM) or TNFα (25 ng/ml) in culture media for 3 h and flow cytometry analysis.
Mass spectrometry
Lipid extraction from purified mitochondria was accomplished as described earlier and extracts were analyzed by nano-electrospray time of flight mass spectrometry using an LCT instrument (Micromass, Elstree, UK) set in positive ion mode.33 A few microliters of the extracts were injected using capillary voltage at 2250 V and sample cone at 35 V. Semi-quantitative evaluation of lipid peaks was undertaken using a set of internal reference ions that maintained equivalent relationships within multiple spectra of the same sample and showed little variation between WT and HtrA2 KO samples. The data were normalized to the major phosphatidyl choline, that is PO-PC (mass-to-charge ratio m/z 758), which was in turn normalized to other reference ions such as C-24 ceramide (m/z 685).34
Fluorescence microscopy
MEFs were plated on 24-well dishes loaded with the superoxide indicator MitoSOX (Invitrogen) at 5 μM in HEPES buffer and then treated with 6-OHDA for 3 h. Cells were then loaded with Hoechst 33342 to stain the cell nucleus (Invitrogen). Live cell microscopy was performed using a KineticScan HCS as described earlier.6
Microarray acquisition and analysis
Experiments using Affymetrix GeneChip MOE430A or Mouse Genome 2.0 (MOE430v2) oligonucleotide arrays were performed using standard Affymetrix protocols. A complete description of all procedures is available online at http://www.affymetrix.com/support/technical/manual/expression_manual.affx and at http://bioinf.picr.man.ac.uk/mbcf/protocols.jsp. Probe level intensities were quantile normalized and quantified using RMA.35 Data analysis was performed with the Genespring software version 3.0 (Silicon Genetics, now Agilent, West Lothian, UK) or Bioconductor software36 for the MEF or cortex brain array, respectively. Probe sets differentially expressed in rotenone-treated MEFs compared with vehicle-treated cells were identified separately in WT and HtrA2 KO MEFs by one-way analysis of variance (ANOVA) using a false discovery rate (FDR) of 0.15. Probe sets differentially expressed in HtrA2 KO cortex tissue compared with WT cortex tissue were identified by one-way ANOVA using a FDR of 0.05, which returned 792 probe sets. Application of a FDR of 0.01 did result in the identification of 79 probe sets differentially expressed in HtrA2 KO cortex tissue. The NCBI GEO accession number for the arrays outlined here is GSE13035.
GSEA
GSEA was performed using GeneSetTest implemented in Bioconductor.36 Gene sets tested were curated from rotenone-induced probe sets in MEFs (this study), the genes induced in MN9D cells by both 6-OHDA or MPP+, and additional genes induced by 6-OHDA treatment,14 as well as the genes induced in an ATF4-dependent manner.18
Respirometry
Mitochondrial respiration was assayed at 37 °C by high-resolution respirometry using an OROBOROS Oxygraph. The DatLab software package (OROBOROS, Innsbruck, Austria) was used for data acquisition (2 s time intervals) and analysis, including calculation of the time derivative of oxygen concentration, signal deconvolution dependent on the response time of the oxygen sensor and correction for instrumental background oxygen flux. Chemical background correction was performed for ascorbate-TMPD auto-oxidation. Complex I activity was assayed in MiR05 respiration buffer (20 mM HEPES, 10 mM KH2PO4, 110 mM sucrose, 20 mM taurine, 60 mM, K-lactobionate, 0.5 mM EGTA, 3 mM MgCl2 × 6H2O, BSA (fatty acid free 1 g/l) in the presence of malate (2 mM), glutamate (10 mM) and ADP (5–10 mM)). Complex II was assayed in respiration buffer supplemented with rotenone (1 μM), succinate (10 mM) and ADP. Complex IV was assayed in respiration buffer supplemented with antimycin A (5 μM) ADP, ascorbate (2 mM), TMPD (0.5 mM) in the presence or absence of exogenous cytochrome c (10 μM). ADP was not added for measurement of respiration in uncoupled mitochondria obtained by one freeze (liquid nitrogen)–thaw cycle of cortex tissue.
Statistical analysis
Data are represented as mean values and the error bars indicate±S.D. or ±S.E.M. as indicated. Groups were compared by two-tailed unpaired t-test, and one- or two-way ANOVA using Bonferroni's post-test using Prism statistical analysis software (GraphPad Software, La Jolla, CA, USA). The significance is indicated as *** for P<0.001, ** for P<0.01 and * for P<0.05.
Accession codes
Abbreviations
- 6-OHDA:
-
6-hydroxydopamine
- DA:
-
dopamine
- ER:
-
endoplasmic reticulum
- GSEA:
-
gene set enrichment analysis
- H2DCF-DA:
-
2′–7′-dichlorofluorescein diacetate
- IPD:
-
idiopathic PD
- ISR:
-
integrated stress response
- KO:
-
knockout
- MEF:
-
mouse embryonic fibroblast
- MPP+:
-
1-methyl-4-phenylpyridinium
- NAC:
-
N-acetyl-L-cysteine
- P:
-
post natal
- PD:
-
Parkinson's disease
- qPCR:
-
reverse transcriptase quantitative real time PCR
- ROS:
-
reactive oxygen species
- UPR:
-
unfolded protein response
- WT:
-
wild type
References
Faccio L, Fusco C, Chen A, Martinotti S, Bonventre JV, Zervos AS . Characterization of a novel human serine protease that has extensive homology to bacterial heat shock endoprotease HtrA and is regulated by kidney ischemia. J Biol Chem 2000; 275: 2581–2588.
Ekert PG, Vaux DL . The mitochondrial death squad: hardened killers or innocent bystanders? Curr Opin Cell Biol 2005; 17: 626–630.
Jones JM, Datta P, Srinivasula SM, Ji W, Gupta S, Zhang Z et al. Loss of Omi mitochondrial protease activity causes the neuromuscular disorder of mnd2 mutant mice. Nature 2003; 425: 721–727.
Martins LM, Morrison A, Klupsch K, Fedele V, Moisoi N, Teismann P et al. Neuroprotective role of the Reaper-related serine protease HtrA2/Omi revealed by targeted deletion in mice. Mol Cell Biol 2004; 24: 9848–9862.
Strauss KM, Martins LM, Plun-Favreau H, Marx FP, Kautzmann S, Berg D et al. Loss of function mutations in the gene encoding Omi/HtrA2 in Parkinson's disease. Hum Mol Genet 2005; 14: 2099–2111.
Plun-Favreau H, Klupsch K, Moisoi N, Gandhi S, Kjaer S, Frith D et al. The mitochondrial protease HtrA2 is regulated by Parkinson's disease-associated kinase PINK1. Nat Cell Biol 2007; 9: 1243–1252.
Spiess C, Beil A, Ehrmann M . A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 1999; 97: 339–347.
Walsh NP, Alba BM, Bose B, Gross CA, Sauer RT . OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell 2003; 113: 61–71.
Lin MT, Beal MF . Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 2006; 443: 787–795.
Ron D, Walter P . Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 2007; 8: 519–529.
Haynes CM, Petrova K, Benedetti C, Yang Y, Ron D . ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev Cell 2007; 13: 467–480.
Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ . A mitochondrial specific stress response in mammalian cells. EMBO J 2002; 21: 4411–4419.
Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 2000; 6: 1099–1108.
Holtz WA, O’Malley KL . Parkinsonian mimetics induce aspects of unfolded protein response in death of dopaminergic neurons. J Biol Chem 2003; 278: 19367–19377.
Ryu EJ, Harding HP, Angelastro JM, Vitolo OV, Ron D, Greene LA . Endoplasmic reticulum stress and the unfolded protein response in cellular models of Parkinson's disease. J Neurosci 2002; 22: 10690–10698.
Braak H, Del Tredici K, Rub U, de Vos RA, Jansen Steur EN, Braak E . Staging of brain pathology related to sporadic Parkinson's disease. Neurobiol Aging 2003; 24: 197–211.
Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003; 34: 267–273.
Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 2003; 11: 619–633.
Silva RM, Ries V, Oo TF, Yarygina O, Jackson-Lewis V, Ryu EJ et al. CHOP/GADD153 is a mediator of apoptotic death in substantia nigra dopamine neurons in an in vivo neurotoxin model of parkinsonism. J Neurochem 2005; 95: 974–986.
Martins LM, Iaccarino I, Tenev T, Gschmeissner S, Totty NF, Lemoine NR et al. The serine protease Omi/HtrA2 regulates apoptosis by binding XIAP through a reaper-like motif. J Biol Chem 2002; 277: 439–444.
Sklar JG, Wu T, Kahne D, Silhavy TJ . Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev 2007; 21: 2473–2484.
Cristea IM, Degli Esposti M . Membrane lipids and cell death: an overview. Chem Phys Lipids 2004; 129: 133–160.
Carriere A, Carmona MC, Fernandez Y, Rigoulet M, Wenger RH, Penicaud L et al. Mitochondrial reactive oxygen species control the transcription factor CHOP-10/GADD153 and adipocyte differentiation: a mechanism for hypoxia-dependent effect. J Biol Chem 2004; 279: 40462–40469.
Holtz WA, Turetzky JM, Jong YJ, O’Malley KL . Oxidative stress-triggered unfolded protein response is upstream of intrinsic cell death evoked by parkinsonian mimetics. J Neurochem 2006; 99: 54–69.
Ruiz N, Silhavy TJ . Sensing external stress: watchdogs of the Escherichia coli cell envelope. Curr Opin Microbiol 2005; 8: 122–126.
Dexter D, Carter C, Agid F, Agid Y, Lees AJ, Jenner P et al. Lipid peroxidation as cause of nigral cell death in Parkinson's disease. Lancet 1986; 2: 639–640.
Guyton KZ, Xu Q, Holbrook NJ . Induction of the mammalian stress response gene GADD153 by oxidative stress: role of AP-1 element. Biochem J 1996; 314 (Part 2): 547–554.
Mengesdorf T, Althausen S, Paschen W . Genes associated with pro-apoptotic and protective mechanisms are affected differently on exposure of neuronal cell cultures to arsenite. No indication for endoplasmic reticulum stress despite activation of grp78 and gadd153 expression. Brain Res Mol Brain Res 2002; 104: 227–239.
Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B et al. IRE1 signaling affects cell fate during the unfolded protein response. Science 2007; 318: 944–949.
Tajiri S, Oyadomari S, Yano S, Morioka M, Gotoh T, Hamada JI et al. Ischemia-induced neuronal cell death is mediated by the endoplasmic reticulum stress pathway involving CHOP. Cell Death Differ 2004; 11: 403–415.
Pennuto M, Tinelli E, Malaguti M, Del Carro U, D’Antonio M, Ron D et al. Ablation of the UPR-mediator CHOP restores motor function and reduces demyelination in Charcot–Marie–Tooth 1B mice. Neuron 2008; 57: 393–405.
Chao JR, Parganas E, Boyd K, Hong CY, Opferman JT, Ihle JN . Hax1-mediated processing of HtrA2 by Parl allows survival of lymphocytes and neurons. Nature 2008; 452: 98–102.
Sorice M, Circella A, Cristea IM, Garofalo T, Di Renzo L, Alessandri C et al. Cardiolipin and its metabolites move from mitochondria to other cellular membranes during death receptor-mediated apoptosis. Cell Death Differ 2004; 11: 1133–1145.
Sandra F, Degli Esposti M, Ndebele K, Gona P, Knight D, Rosenquist M et al. Tumor necrosis factor-related apoptosis-inducing ligand alters mitochondrial membrane lipids. Cancer Res 2005; 65: 8286–8297.
Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP . Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 2003; 31: e15.
Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004; 5: R80.
Acknowledgements
We thank Darren Moore, Anne Bertolotti and Emma Deas for technical advice. Valina Dawson is thanked for the generous gift of anti-DJ-1 and David Knight for his help with mass spectrometric analysis. We also thank Pierluigi Nicotera for advice. K Klupsch was funded by a PhD studentship from Boehringer Ingelheim Fonds.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Edited by G Kroemer
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Supplementary information
Rights and permissions
About this article
Cite this article
Moisoi, N., Klupsch, K., Fedele, V. et al. Mitochondrial dysfunction triggered by loss of HtrA2 results in the activation of a brain-specific transcriptional stress response. Cell Death Differ 16, 449–464 (2009). https://doi.org/10.1038/cdd.2008.166
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2008.166
Keywords
This article is cited by
-
Signaling pathways in Parkinson’s disease: molecular mechanisms and therapeutic interventions
Signal Transduction and Targeted Therapy (2023)
-
Protease-independent control of parthanatos by HtrA2/Omi
Cellular and Molecular Life Sciences (2023)
-
Evaluation of animal model congruence to human depression based on large-scale gene expression patterns of the CNS
Scientific Reports (2022)
-
Mitochondrial quality control: from molecule to organelle
Cellular and Molecular Life Sciences (2021)
-
The oligosaccharide portion of ganglioside GM1 regulates mitochondrial function in neuroblastoma cells
Glycoconjugate Journal (2020)