Abstract
Base excision repair (BER) is an evolutionarily conserved process for maintaining genomic integrity by eliminating several dozen damaged (oxidized or alkylated) or inappropriate bases that are generated endogenously or induced by genotoxicants, predominantly, reactive oxygen species (ROS). BER involves 4-5 steps starting with base excision by a DNA glycosylase, followed by a common pathway usually involving an AP-endonuclease (APE) to generate 3′ OH terminus at the damage site, followed by repair synthesis with a DNA polymerase and nick sealing by a DNA ligase. This pathway is also responsible for repairing DNA single-strand breaks with blocked termini directly generated by ROS. Nearly all glycosylases, far fewer than their substrate lesions particularly for oxidized bases, have broad and overlapping substrate range, and could serve as back-up enzymes in vivo. In contrast, mammalian cells encode only one APE, APE1, unlike two APEs in lower organisms. In spite of overall similarity, BER with distinct subpathways in the mammals is more complex than in E. coli. The glycosylases form complexes with downstream proteins to carry out efficient repair via distinct subpathways one of which, responsible for repair of strand breaks with 3′ phosphate termini generated by the NEIL family glycosylases or by ROS, requires the phosphatase activity of polynucleotide kinase instead of APE1. Different complexes may utilize distinct DNA polymerases and ligases. Mammalian glycosylases have nonconserved extensions at one of the termini, dispensable for enzymatic activity but needed for interaction with other BER and non-BER proteins for complex formation and organelle targeting. The mammalian enzymes are sometimes covalently modified which may affect activity and complex formation. The focus of this review is on the early steps in mammalian BER for oxidized damage.
Similar content being viewed by others
Introduction
The genome is inherently unstable due to spontaneous chemical reactions, and its fidelity is compromised due to very low but significant replication errors. Moreover, the genomes of all organisms are continuously exposed to a wide variety of insults, e.g., endogenous reactive oxygen species (ROS), and environmental genotoxic agents such as cigarette smoke, UV light, and ionizing radiation. Furthermore, chemotherapeutic drugs including alkylating agents are invariably genotoxic and induce damage in both healthy and tumor cell genomes. ROS, because of their continuous generation as by-products of respiration in the mitochondria 1 represent the predominant group of chemicals that damage genomes of aerobic organisms. ROS are also generated by ionizing radiation and during oxidative metabolism of xenobiotic agents by p450s. ROS include O2•- (superoxide radical anion), OH• (hydroxyl radical) and H2O2 (hydrogen peroxide). O2•- is also generated during inflammatory response by NADH oxidases 2. ROS-induced cellular changes have been implicated in a multitude of diseases, ranging from cardiovascular dysfunction to arthritis and cancer, as well as in aging and age-related disorders. To counteract ROS-induced DNA damage, cells have evolved several defense mechanisms that act at different levels to prevent or repair such damage.
The base excision repair (BER) pathway is responsible for repairing most endogenous base lesions and abnormal bases in the genome as well as similar lesions generated by several groups of environmental agents, or their metabolic intermediates. The BER pathway is also involved in repair of DNA single–strand breaks. Such breaks resulting from free radical reaction of deoxyribose residues invariably possess blocked termini. Contrary to the earlier concept that BER is a simple process requiring very few proteins, recent discoveries paint a more complex picture of mammalian BER with several subpathways. This review is exclusively focused on the early steps in BER, in particular, for repair of the most predominant damage namely, those induced by ROS. The review of Horton et al. in this issue is devoted to some aspects of the late steps in BER.
DNA repair: A recipe for survival
It has been generally estimated that some 104 lesions are induced in a mammalian cell genome every day by spontaneous chemical reactions and during cellular metabolism. The most predominant among these are spontaneously generated or products of ROS reaction 3. Environmental chemicals, tobacco smoke and radiation add to the damage load. Most ROS-induced oxidized base lesions and abasic (AP) sites if left alone could be replicated by replicative or DNA translesion synthesis (TLS) polymerases 4, 5. Misreplication of oxidized bases and noninstructional AP sites would often give rise to point and sometimes more complex mutations. Single nucleotide polymorphisms (SNPs), observed at quite high frequency (1 in 300 bp) in mammalian genomes, likely resulted from such spontaneous mutations. The origin of sporadic cancer is also very likely due to such spontaneous mutations in oncogenes and tumor suppressor genes 6. For example, C is spontaneously deaminated to U and ROS could oxidatively deaminate C to 5-hydroxyuracil (5-OHU). Both U and 5-OHU preferably pair with A during DNA replication by replication or TLS DNA polymerases resulting in GC→AT transition mutation. Similarly, G is oxidized to 8-oxo-7,8 dihydroguanine (8-oxoG) by a variety of ROS 7. Its mispairing with A would give rise to GC→TA transversion mutation 8. Such mutations have indeed been preferentially observed in cellular genomes after oxidative stress 9. Defense mechanisms to maintain genomic integrity in the face of continuous assault on the genome evolved in all organisms primarily in the form efficient DNA repair. Thus, repair of oxidized base lesions is essential for maintaining genome integrity and survival.
Base excision repair (BER): A pathway for processing small base adducts, inappropriate or oxidized bases, and DNA single-strand breaks
BER is the most versatile among excision repair pathways, and is responsible for repairing most endogenous lesions like oxidized bases and AP sites, as well as DNA single-strand breaks. The basic mechanism of BER was first elucidated in E. coli 10. Subsequent studies showed that the process is conserved in the eukaryotes including mammals. Until recently, BER was believed to be the simplest, thoroughly characterized process among all repair pathways. Although BER is responsible for repair of a majority of cellular DNA damages 3, 11, the functional significance of BER in prevention of disease remains unclear. This is presumably because nucleotide excision repair (NER) and DNA mismatch repair (MMR) deficiencies have been linked to cancer and other diseases, whereas no disease phenotype has been linked to BER deficiency so far 12.
In addition to oxidation of DNA bases, ROS attack deoxyribose in DNA to generate strand breaks with nonligatable dirty ends (sugar fragments or 3′ phosphates). These are also repaired by the BER pathway. The 3′ blocking groups include 3′ phosphate, 3′ phosphoglycolaldehyde, or 3′ phosphoglycolate 13. The 5′ terminus normally contains phosphate but after ROS reaction the nonligatable ends include 5′ OH and 5′ phosphodeoxyribose derivatives such as 2-deoxyribonolactone 14.
Basic mechanism of BER
BER requires only four or five enzymes in the basic reaction steps to carry out repair of DNA containing AP sites or base damage. These include a DNA glycosylase, an AP endonuclease, a DNA polymerase, and a DNA ligase 15. BER is initiated with excision of a damaged base by the DNA glycosylase. An AP site is generated due to cleavage of the N-glycosidic bond of the damaged base. The APE cleaves the AP site in the second step, to generate 3′ OH and 5′ deoxyribose phosphate (dRP) terminus 16. The third reaction in the pathway involves the DNA polymerase to fill in the single nucleotide gap generated due to lesion base removal. In mammalian cells, the repair DNA polymerase β (Polβ) has an intrinsic dRP lyase activity 17, 18 which cleaves the dRP residue to generate 5′ phosphate and the resulting nick after single nucleotide incorporation is sealed by the DNA ligase in the final step. These BER enzymes are conserved from E. coli to mammals.
DNA glycosylase, a key enzyme in BER
BER is unique among the excision repair processes in that the individual base lesions are recognized by distinct DNA glycosylases less than a dozen of which are expressed in E. coli or mammalian cells. However, each DNA glycosylase acts on a limited number of damaged bases. DNA glycosylases are relatively small (∼30-50 kDa) monomeric proteins that do not require cofactors for their activity. These enzymes generally excise altered bases that cause only minor perturbations in the DNA helix. Table 1 provides the list of DNA glycosylases characterized in E. coli and mammals so far. The first DNA glycosylase to be discovered was uracil DNA glycosylase (UDG) in E. coli after searching for an enzymatic activity that would recognize uracil whose presence in DNA as a G•U pair would be mutagenic because U is generated due to deamination of cytosine 19. Subsequently, similar enzyme activities were identified in other bacteria, yeast, plants and mammalian cells, and mitochondria-specific UDGs were also characterized 20. In contrast to the promiscuity of most DNA glycosylases for diverse substrates, UDG is highly specific for U which tightly fits into UDG's catalytic pocket. Elucidation of the crystal structures and mutational studies of UDG 21 have established the structural basis for substrate specificity in preference to structurally similar normal bases like C or T in DNA. DNA binds along a positively charged groove in the enzyme, but the tight-fitting uracil-binding pocket located at the base of this groove is too deep and narrow to allow binding of DNA-uracil unless it is 'flipped out' of the DNA helix. The structure of human UDG and that bound to uracil-containing oligo has demonstrated the basis for the enzyme-assisted nucleotide flipping 21, 22. Another DNA glycosylase SMUG1 has been characterized in human cells 23, 24, which catalyzes excision of U and also oxidized pyrimidines such as 5-hydroxycytosine (5-OHC). SMUG1 is a member of UDG family of glycosylases. Finally, a third DNA glycosylase in mammalian cells, thymine-(T•G)-DNA glycosylase (TDG) was shown to excise T and U from their base pairs with G, and may be involved in repairing T generated by deamination of 5-methyl C in a methylated C•G pair 25.
As discussed later, DNA glycosylases specific for oxidized base lesions are bifunctional with intrinsic AP lyase activity that causes cleavage of DNA strand at the AP site generated after base excision 26. In this case, the 3′ terminus at the cleavage site is nonligatable and blocked with α,β-unsaturated aldehyde dehydration product of deoxyribose phosphate or only phosphate, and the 5′ terminus contains a phosphate residue (Figures 1 and 2). Unlike in the case of APE cleavage product of an AP site where the 5′ terminus is blocked with deoxyribose phosphate, the 3′ terminus of ROS-induced DNA strand breaks is blocked, and needs to be cleaned prior to the subsequent step in BER.
A schematic illustration of BER subpathways for damaged bases and DNA strand breaks. The damaged base is represented as a star (*). Divergent base excision steps converge to common steps for end processing, followed by repair DNA synthesis (represented as blue dots) and strand sealing. Polβ could also be involved in LP-BER by collaborating with FEN-1. Other details are discussed in the text.
Repair of alkylated bases by monofunctional DNA glycosylases and by MGMT
Endogenous base lesions also include methyl adducts which are likely to be generated by chemical reactions with S-adenosylmethionine 27. The methylated base adducts like other base lesions could also be generated by reactions with exogenous methylating agents including many chemotherapeutic drugs and N-methylnitrosamines 28. A multitude of methyl base adducts are produced many of which including O6-methylguanine and O4-methylthymine are highly mutagenic. The methylated adducts are repaired in all organisms via two pathways. One is a direct reversal of damage without DNA repair synthesis. This was first discovered in E. coli as an adaptive response and the protein named Ada 29 turned out to be conserved in eukaryotes and mammals and was named O6-methylguanine-DNA methyltransferase (MGMT) 27. MGMT demethylates O6-methylG in situ in DNA and also O4-methylT. A second direct reversal enzyme named AlkB in E. coli and ABH in mammals demethylates 1-methylA and 3-methylC with more complicated reaction chemistry 30. In contrast, other methyladducts as well as hypoxanthine and xanthine, deamination products of A and G respectively, are repaired via the conventional BER pathway by N-methylpurine-DNA glycosylase (MPG, also named MAG). This enzyme and its E. coli ortholog AlkA excise N-alkylpurines, primarily 3-methylA, 3-methylG and 7-methylA. E. coli expresses another glycosylase Tag which specifically excises 3-methyladenine. AlkA also excises O2-methyladducts of C and T 31, 32.
DNA glycosylases for normal bases
While DNA glycosylases in general recognize only abnormal bases including U in DNA as substrates, there is one exception for mismatch-specific glycosylase MutY. MutY was discovered in E. coli as a mutator gene whose deficiency induces spontaneous C•G→A•T transversion mutation. It excises primarily the normal base A (and also G) from A•G and A(G)•8-oxoG base pair mismatches 33, 34. Thus MutY and its mammalian homolog MYH apparently serve an important function in protection against oxidative damage by removing the base A incorporated in the nascent strand opposite 8-oxoG during DNA replication 9, 35, 36.
Bifunctional DNA glycosylases
The monofunctional glycosylases excise the substrate base, leaving an intact AP site. In contrast, the bifunctional glycosylases have an additional lyase activity. This second intrinsic activity is invariably present in DNA glycosylases specific for oxidized bases. The monofunctional DNA glycosylases generate an AP site which is processed by an AP endonuclease. Bifunctional DNA glycosylases further process the AP site via β or βδ elimination reaction. A mechanistic distinction between these two types of enzymes is that monofunctional DNA glycosylases typically use an activated water molecule as a nucleophile in attacking sugar C1′ of the target nucleotide, where bifunctional glycosylases/AP lyases often use ɛ-NH2 of a lysine or the N-terminal proline as the active site nucleophile 37. An intermediate step is the formation of a transient Schiff base between the amino group and the C1′ of deoxyribose for both base excision and DNA strand cleavage. Usually, the base excision and lyase reaction steps act in a concerted sequence. However, in some cases, e.g., for 8-oxoguanine-DNA glycosylase (OGG1), the lyase reaction is very weak. Thus, intact AP sites are the major product after OGG1-catalyzed cleavage of 8-oxoguanine from DNA 38, 39.
Despite differences in the specific residues used to recognize damaged bases, one unifying mechanism for BER initiation by DNA glycosylases is the extrahelical flipping of the damaged deoxynucleotide into a lesion-specific recognition pocket. All DNA glycosylases studied so far bind to the minor groove, kink DNA at the site of damage, and flip the lesion base out of the DNA major groove. Thus each DNA glycosylase is damage specific, and only those bases that can be accommodated in the defined binding pocket after nucleotide flipping provide the necessary contacts and orientation for their excision. The initial recognition apparently exploits the deformability of the DNA at a base pair destabilized by the presence of a lesion 40. It is likely that plasticity of the catalytic pocket allows induced fit of diverse substrates. This could also explain why DNA glycosylases (with the exception of UDG) are poor enzymes with low turnover.
Oxidized base-specific DNA glycosylases
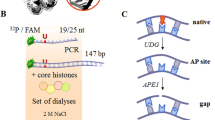
Oxidized base-specific DNA glycosylases were also first discovered in E. coli, along with subsequently characterized eukaryotic orthologs. They are categorized into two families based on tertiary structure, active site characteristics and AP lyase reaction, and are named after the prototype Nth [endonuclease III] and Fpg (formamidopyrimidine-DNA glycosylase) 37. The Nth family utilizes an internal Lys residue as the active site for β elimination reaction, generating a 3′ phospho α,β-unsaturated aldehyde (3′ PUA), also named 3′ phosphor 4-hydroxylpentenal, at the strand break. In contrast, the members of the E. coli Fpg family including Nei (endonuclease VIII) catalyze βδ elimination at the AP site by utilizing N-terminal Pro as the nucleophile and remove the deoxyribose residue to produce a 3′ phosphate terminus at the DNA strand break 41. In both cases, the 5′ terminus retains the phosphate moiety. Figure 1 depicts the β and βδ elimination reactions.
DNA glycosylases could also be classified into two distinct families based on tertiary structural features, namely, the presence of either an HhH or an H2TH motif unrelated to the type of oxidized base recognized 40. Representative structures have been determined for both families. However, some members of these families have common substrates. OGG1, MutY, Nth and their homologs have HhH structure 42, 43, 44; while MutM (Fpg) and Nei have H2TH motif 44, 45, 46, 47.
Mammalian DNA glycosylases for oxidized bases
Four oxidized base-specific glycosylases have been characterized in mammalian cells. NTH1 and OGG1 were characterized previously, both of which belong to the Nth family 38, 48, 49. Thus, both NTH1, preferring oxidized pyrimidines as substrates, and OGG1, primarily responsible for the removal of 8-oxoG and ring opened guanine, i.e., formamidopyrimidine (Fapy-G), catalyze β elimination 11, 49. Subsequently, we characterized two additional human DNA glycosylases (also independently discovered by others), which belong to the Fpg/Nei family; we named these NEIL1 and NEIL2 50, 51, 52, 53, 54, 55. All these enzymes as already indicated are bifunctional glycosylases with broad substrate range. We also identified a third member of this family, namely NEIL3, but no glycosylase activity has been characterized for this enzyme so far (Izumi T, unpublished data).
Among the three NEILs, N-terminal Pro, present only in NEIL1 and NEIL2, acts as the active site nucleophile 50, 53. NEILs are closer to Nei than Fpg, and we and others initially showed that both NEIL enzymes prefer modified pyrimidine substrates 53. However, NEIL1 was later shown to prefer ring-opened purines, i.e., formamidopyrimidine (Fapy)-A and -G, which along with 8-oxoG are the preferred substrates for E. coli Fpg. While we initially showed 8-oxoG as a poor substrate of NEILs on duplex DNA substrate, others have shown NEIL1's robust 8-oxoG excision activity 54, 56. Subsequently, we showed that NEILs' activity and substrate specificity depend largely on the DNA structure, and NEILs have significant 5-hydroxyuracil excision activity towards single-stranded or bubble DNA 57. Interestingly there is no evidence for NEILs in the yeast which however, has one OGG1 and two distinct NTHs (Ntg) 58.
Table 2 lists the comparative characteristics of OGG1, NTH1, NEIL1, and NEIL2 all of which have substrate preference but not absolute specificity.
Nonessentiality of individual DNA glycosylases in mammals
The genes and cDNAs of mammalian OGG1 and NTH1 were cloned in late 1990s, and NEIL1 and NEIL2 genes were identified in mammalian genome databases in 2002. The genomic cloning led to the generation of mouse mutants lacking OGG1, NTH1 and NEIL1 as well as MYH 59, 60, 61. All of the mouse mutants and their embryonic fibroblasts are viable. Furthermore, no strong phenotype was observed in the mutant mice in regard to enhanced incidence of spontaneous cancer or accelerated aging. Although a small increase in cancer incidence was observed in OGG1-null mice after exposure to bromate 62, the lack of immediate effect was unexpected, particularly because the deficiency of other BER proteins, such as APE1 and DNA Polβ, results in embryonic lethality in mutant mice 63, 64. There is compelling evidence that the oxidized base lesions are endogenously generated at a reasonably high frequency many of which are mutagenic and thus potentially carcinogenic. Repair of such mutagenic and carcinogenic lesions is essential, which is presumably carried out by other glycosylases in the cells deficient in a single enzyme. Furthermore, enhanced accumulation of 8-oxoG, observed in DNA isolated from various tissues of OGG1 null mice, confirms the requirement of OGG1 in removing such lesions. In contrast, deficiency in NER or MMR processes significantly enhances cancer susceptibility 65, 66, 67. One possible explanation for the lack of phenotype of the glycosylase mutants is that, unlike in NER or MMR, multiple glycosylases could provide back-up functions because of their overlapping substrate range. We could argue that oxidative damage repair is too important to be left to a single repair enzyme. At the same time, as we have already pointed out, the glycosylases do have preferred substrates and it is also likely that the individual enzyme acts preferentially depending on the state of the genome, i.e., whether the cells are cycling or postmitotic and whether the damage is located in transcriptionally active vs. inactive sequences 68. This could also explain why accumulation of oxidized bases in the genomes of OGG1 or NTH1 null mouse cells does not cause any obvious phenotype 69. Although oxidized bases could possibly be repaired alternatively via the NER pathway, this is unlikely to be a major alternative repair mode for most oxidized bases 70, because such bases do not induce significant perturbations in the duplex DNA structure needed for recognition by the NER proteins. We have proposed a hypothesis that the lesions in transcriptionally active sequences are preferentially repaired by other glycosylases such as NEILs while OGG1 and NTH1 act globally in repairing both active and inactive genome sequences 71.
More than a dozen oxidatively damaged bases have been identified in the genome but only a few DNA glycosylases have been characterized. This is consistent with the observation that the glycosylases have broad substrate range. These enzymes also have very low turn-over, presumably as the price to pay for their promiscuity. This is especially true for mammalian oxidized base-specific DNA glycosylases. However, these enzymes may also have distinct in vivo functions, although the requirement is not absolute as the deficiency of a given DNA glycosylase is not lethal. Thus the distinction among different BER sub-pathways initiated with different DNA glycosylases is not absolute; the sub-pathways may be preferentially used depending on specific cellular states, location of the damage site in inactive vs. active regions of the genome and the nature of the damage itself.
Unique DNA structure preference of NEILs
There is one absolute distinction in substrate recognition specificity between two families of oxidized base-specific glycosylases. OGG1 and NTH1 excise base lesions only from duplex DNA, which is expected because the undamaged strand provides the template for repair of the damaged strand. So we were greatly surprised to observe that NEIL1 and NEIL2 preferentially excise base lesions from single-stranded DNA, including bubble or forked DNA structures 57. Yasui's group had made a similar observation earlier for NEIL1 55. Because the bubble and fork structures mimic the transcription and replication intermediate respectively, we have proposed preferential involvement of NEILs in repair of oxidized bases during transcription or DNA replication. Our initial observation of S-phase-specific activation of NEIL1 and not of other three glycosylases suggested NEIL1's preferential involvement in repair of base lesions in template DNA prior to replication. We have named this subpathway replication-associated BER (RA-BER) 57. A similar model was earlier proposed for repair of U in replicating DNA by UNG2 which also shows S-phase specific activation 72. Thus SMUG1 may preferentially act in repairing U in noncycling genomes 73.
While the RA-BER model and NEIL1's role therein are not established, our recent observations on the interaction of NEIL1 with the sliding clamp PCNA and DNA damage-inducible alternative clamp 9-1-1 and NEIL1's activation by these proteins support our hypothesis 74, 75.
Two basic modes of BER: SN- vs. LP-BER
In the simple BER model, excision of the damaged base leaves a 1-nt gap whose repair has been named single nucleotide repair (SN-BER). This has also been referred to as short-patch repair (SP-BER) 76. In contrast, a repair patch size of 2-8 deoxynucleotides was observed initially in reconstituted BER systems and subsequently in vivo, which was named long-patch BER (LP-BER). During in vitro repair of an AP site analog, lacking the aldehyde group, Polβ could not remove the APE-generated 5′ blocking group via its dRP lyase activity 77, 78, 79, 80. Matsumoto's and Dogliotti's groups first showed that such a 5′ blocking group could be removed by the 5′-flap endonuclease 1 (FEN-1) normally required for removing the 5′ RNA primers from Okazaki fragments during DNA replication. Other steps of LP-BER are identical to that of DNA replication involving DNA polymerases δ/ɛ and DNA ligase I (Lig I). These enzymes including FEN-1 are recruited by the sliding clamp PCNA which enhances their activity 77, 78, 79. The choice of LP-BER vs. SN-BER may thus depend on, among other factors, the state of the 5′-dRP terminal moiety. With unaltered aldehyde group in deoxyribose Polβ could carry out SN-BER by excising the 5′-dRP 17, 81. On the other hand, LP-BER is necessary when the AP sites are further oxidized by ROS, and the resulting 5′ blocking groups after cleavage of the DNA strand by APE could not be removed by Polβ's 5′ end cleaning lyase activity. The LP-BER subpathway utilizes the DNA replication machinery where FEN-1 excises the 5′-oxidized dRP-containing segment displaced as a flap during repair 82. More recently, FEN-1 was found to have additional 5′ exonuclease and gap-specific endonuclease activities 83. The role of FEN-1's exonuclease function in LP-BER however, is yet to be clarified. PCNA, loaded on duplex DNA by replication factor-C (RFC), acts as a sliding clamp in LP-BER as it does during replication 84. It is likely that Polδ/ɛ can substitute for Polβ in repair synthesis and carry out LP-BER, because Polβ-null cells grow normally and are not hypersensitive to oxidative stress 85. However, Polβ also interacts with FEN-1 and Lig I and could thus carry out LP-BER 86. These distinct pathways are schematically shown in Figure 1.
Repair of AP sites and single-strand interruption with nonligatable termini: a common step in the BER pathway
DNA glycosylases excise damaged or inappropriate bases generating repair intermediates, namely AP sites and strand breaks with blocked termini. Repair of these lesions shares common steps with that of ROS-induced DNA single-strand breaks, and spontaneously generated AP sites. AP sites could be further oxidized to various products with oxidation and fragmentation of deoxyribose moiety and strand cleavage 87. The major products include 2′-deoxyribonolactone, 2-deoxypentos-4-ulose and 3′ phosphoglycolate 14. Unmodified AP sites are also generated by monofunctional DNA glycosylases, as already mentioned. But, all oxidized base-specific glycosylases generate 3′ phospho α,β unsaturated aldehyde or 3′ phosphate respectively, and 5′ phosphate at the strand break. In addition to the AP sites, the single-strand breaks with 3′ blocking residues have to be repaired to maintain genomic integrity.
AP-endonuclease (APE), a ubiquitous repair protein with dual nucleolytic activities
In view of continuous generation of AP sites and DNA strand breaks, all of which are more toxic than damaged bases themselves 88, it is not surprising that multiple enzymatic processes have evolved to repair these lesions among which APE is the most prominent and ubiquitous 88. APE was discovered in E. coli as a 3′ exonuclease/DNA 3′ phosphatase and was named exonuclease III (Xth) 89 and subsequently found to be an AP site-specific endonuclease that generates 3′ OH terminus after cleaving the DNA strand 5′ to the abasic site. It is also active as a 3′ exonuclease/phosphatase and produces 3′ OH terminus at strand breaks with 3′ blocks. Another endonuclease, endonuclease IV (Nfo), was subsequently discovered in E. coli whose activities as AP site-specific endonuclease and 3′ end cleaning phosphodiesterase are very similar to that of Xth except that Nfo lacks 3′ exonuclease activity and does not require Mg2+ for catalysis 90, 91. Subsequently APEs were characterized in a variety of organisms ranging from bacteria to yeast, C. elegans and mammals. These enzymes have similar but not identical enzymatic functions and conserved structures, and could be classified as members of the Xth or Nfo family.
Multiple APEs are also present in lower eukaryotes
The in vivo function of E. coli Nfo is not clear because its deletion in cells containing wild type Xth does not cause any phenotype. However, Xth null cells are highly sensitive to ROS confirming that the AP sites and strand breaks with 3′ blocked ends are toxic 89. E. coli lacking both Xth and Nfo are highly sensitive to ROS and alkylating agents, as expected 92. However, the fact that the cells are viable indicates either that AP sites and nonligatable strand breaks could be bypassed or that these are repaired via an alternative back-up mechanism. Noninstructional AP sites are highly mutagenic 93. However, these could be bypassed by translesion synthesis (TLS) DNA polymerases in the absence of repair 94, 95.
There is no evidence so far that oxidized AP sites or strand breaks could be bypassed in vivo. However, an alternative process for repairing these lesions in multi-allelic E. coli or post replication repair could bypass AP sites by strand switching 10, 96, or the AP sites could also be repaired via NER 97. The observation that Xth/Nfo deficient E. coli loses viability when UvrB, a key NER protein, is also inactivated, supports this scenario 92.
Xth and Nfo type APEs, named APN2 and APN1, respectively, are expressed in yeast 98, 99. In contrast to the situation in E. coli, APN1 is the major contributor of APE activity in this unicellular eukaryote. However, as in E. coli, deletion of both APN genes does not cause lethality although it increases sensitivity to alkylating and oxidizing agents. However, simultaneous inactivation of the nucleotide excision repair pathway is lethal as in E. coli 100.
Two distinct types of APE possess nearly identical activity
The structures of E. coli Xth and its mammalian ortholog APE1 as well as those of Xth or Nfo of various microorganisms have been solved by X-ray crystallography 101. It is remarkable that Xth and Nfo have nearly identical enzymatic activity and yet do not have any structural similarity. Xth is a Mg2+-dependent nuclease belonging to the superfamily of pancreatic DNase. The divalent metal bound forms of Xth and human APE1 show one metal ion bound per monomeric protein molecule 102. However, recent molecular dynamic simulation predicts that APE1 does have two potential Mg2+-binding sites A and B, although only one Mg2+ is bound to the protein which shuttles between the two sites during catalysis 103. Mol et al. 102 solved the structure of human APE1 bound to its cleavage product and Mg2+ was shown to be present at the A site. The structure of substrate bound active APE1 could not be solved in the presence of Mg2+ for obvious reasons. The key residues and motifs required for catalytic activity and metal binding in all Xth family members are conserved which allowed our lab to clone the S. cerevisiae Apn2 99. We subsequently identified and cloned S. pombe APN2 based on sequence homology 104.
The Nfo type APE is extremely unusual because it is a Zn metalloenzyme with a 3-Zn2+ center 105. It does not require external Mg2+ for activity 92. The evolutionary significance of the Nfo family is unclear.
Mammalian cells express only Xth type APE, APE1
Unlike in E. coli or yeast, there is no evidence for an Nfo ortholog in mammalian cells. Mammalian APE1 was characterized based on activity more than two decades ago 106. Subsequent to its cloning and extensive characterization of APE1, a second Xth ortholog, named APE2 was cloned 107, 108. However, the recombinant APE2 has negligible APE-specific activity, which suggests its lack of function in BER (108; Izumi T, unpublished data). Nevertheless, expression of APE2 in various mammalian cells does implicate it in some other in vivo functions.
Mammalian APE1 and E. coli Xth have distinct properties
In spite of the similarity in tertiary structure and conserved motifs and catalytic mechanism, mammalian APE1 has many distinctive features starting with its unique N-terminal extension of about 61 residues which are absent in Xth and dispensable for APE activity 109. This segment contains the nuclear localization signal (NLS) 110 and is also required for the transcriptional regulation function unique to APE1 and unrelated to BER 111, 112. We recently discovered that human APE1 (hAPE1) is acetylated in vivo at Lys 6 / Lys 7 which modulates its regulatory but not endonuclease activity 113. The endonuclease and 3′ end cleaning phosphodiesterase activity of APE1 is weaker than that of Xth, but more importantly, APE1 has barely detectable DNA 3′ phosphatase activity and has a very limited 3′ exonuclease activity 114, 115. However, 3′ mismatch-specific exonuclease activity has been observed in APE1 although its in vivo significance has not been established 116.
APE1 is essential in mammalian cells
The attempts of several groups to generate APE1-null mice failed because of very early (4-5 days post coitus) embryonic death. This strongly suggested that unlike in lower organisms, APE1 is essential at least for early embryonic development 64, 117. Efforts to generate cell lines from APE1 null embryos were also uniformly unsuccessful which distinguishes it from Polβ. Null mutation for Polβ in mice is also embryonic lethal (although the embryos survived longer) 118. However, Polβ null mouse embryonic fibroblast lines could be established which have normal viability 119.
We tested whether APE1 is essential in the postembryonic stage by introducing “floxed” hAPE1 transgene in the genome of APE1 heterozygous mice. Our attempt to generate viable mouse embryos lacking endogenous APE1 but expressing ectopic hAPE1, by inbreeding the heterozygous, transgenic mice was unsuccessful. However, we were able to establish embryonic fibroblast lines from such crosses which lack the endogenous APE1 allele but have normal viability. Transgenic expression of Cre in these cells leading to deletion of the APE1 transgene triggered apoptosis. This observation and an independent study provide unequivocal evidence that APE1 is essential even for somatic cells 120, 121. Kelley and his colleagues had earlier shown that APE1 downregulation triggers apoptosis in several human cell lines 122. Further studies showed that both DNA repair and transcriptional regulation activity of APE1 are independently required for cell viability 120.
Accumulation of single-strand breaks in the genome of APE1-null cells
Essentiality of APE1 in mammalian cells but not in lower organisms suggests that some cytotoxic lesions normally repaired via APE1-dependent BER could be alternatively repaired by NER or other pathways in E. coli or yeast. We suggest that these include DNA strand breaks with phosphosugar fragment-containing 3′ termini which are efficiently repaired by the APE1-dependent pathway (Figure 1). We had shown earlier that APE1 activity is limiting in repair of ROS-induced strand breaks but not of AP sites 123. Using embryonic fibroblast line with conditional null mutation of APE1, we observed that APE1 inactivation led to significant increase in single-strand breaks as well as AP sites 112, 124.
A unique PNK-dependent BER subpathway for mammalian cells
NTH1 and OGG1 generate 3′ PUA by β-elimination of deoxyribose phosphate which is efficiently removed by APE1 in the next step. NEILs, on the other hand, have βδ lyase activity and thus generate 3′ phosphate, a poor substrate for APE1. However, mammals, unlike E. coli, express high level of polynucleotide kinase (PNK) with dual 5′ kinase/3′phosphatase activities 125. PNK has already been implicated in repair of ROS-induced single-strand breaks 126. We recently showed that NEIL-initiated BER in mammals unlike in E. coli does not involve APE, but requires PNK 51, 115. The presence of a NEIL-initiated, APE-independent BER subpathway in mammalian cells was further supported by the presence of PNK and not APE1 in the NEIL1 or NEIL2 immunocomplex isolated from cell extracts 51, 115. We also showed that NEILs carry out SN-BER mediated by PNK, Polβ, Lig IIIα and XRCC1. Stable interaction of NEILs with the downstream repair proteins Polβ, Lig IIIα and XRCC1 led us to propose that these DNA glycosylases, as the first enzyme in the repair process, determine the specific BER sub-pathway 51, 115. Thus for oxidized bases, DNA glycosylases/ AP lyases that carry out either β elimination or βδ elimination determine the subsequent steps. APE1 is responsible for processing the β elimination product while PNK is required for generating 3′-OH terminus from 3′-phosphate, a product of βδ elimination catalyzed by NEIL1 or NEIL2. Furthermore, AP sites and 3′ PUA generated by other DNA glycosylases can also be processed through a NEIL-PNK dependent pathway 127. This alternative route of repair thus may provide important redundancy in mammalian BER, a critical safeguard against oxidative and spontaneous DNA damage 71.
Common mechanisms of BER and single-strand break repair: End cleaning at strand breaks
The importance of end cleaning at a strand break as a vital step in BER was recently highlighted 115. Irrespective of the type, the intermediate step in BER is a 'strand break with either 3′ or 5′ or both ends blocked' by residues that are not compatible for subsequent gap filling and ligation. The normal substrate for a DNA polymerase is a gap at a strand break with 3′ OH and 5′ phosphate residues. The common pathways for providing such a repair substrate are as follows: (i) Removal of a base lesion by a monofunctional glycosylase, followed by the action of APE on the resulting AP site would generate a strand break with 3′-OH and 5′ dRP group. Though unmodified 5′ dRP moiety in mammalian cells is removed by Polβ's intrinsic dRP lyase activity, nuclear replicative DNA polymerases Polδ/ɛ lack this activity. Repair synthesis by these enzymes follows the LP-BER subpathway, where the 5′-dRP-containing moiety and 4-6 additional deoxynucleotides are displaced as a single-stranded DNA flap, which is then cleaved by FEN-1. Furthermore, the AP site could be oxidized by ROS and its cleavage by APE would generate 5′ blocking groups which have to be processed by FEN-1 via the LP-BER pathway. (ii) The action of a bifunctional glycosylase with intrinsic β lyase activity would result in a strand break with 3′ PUA, a blocking residue, and 5′ phosphate. The 3′ PUA is removed by the 3′ phosphodiesterase activity of APE1 to generate 3′ OH. (iii) The bifunctional glycosylase with an intrinsic βδ elimination property would generate 3′ phosphate and 5′ phosphate at the strand break. The 3′ end cleaning enzyme here is PNK instead of APE1 as already discussed (Figs. 1, 2).
In addition, the 3′- or 5′- termini of most if not all DNA single-strand breaks directly induced by ROS or radiation are 'blocked' or 'damaged' and must be restored to the conventional 3′-OH and 5′-phosphate moieties in order for gap filling and subsequent ligation to occur 128. The common blocks at ROS induced strand breaks are 3′ phosphoglycolate and 3′ phosphoglycolaldehyde, which are processed by APE1 or 3′ phosphate which is removed by PNK. However, 'end cleaning' is perhaps the most diverse enzymatic step in strand break repair, due to the variety of termini generated. As mentioned earlier, Polβ (5′ dRP lyase) and FEN-1 are used for cleaning the 5′ blocked termini.
Eukaryotic cells express other DNA end cleaning enzymes
Tyrosyl phosphodiesterase 1 (TDP1) has been implicated in end processing at DNA double-strand breaks in budding yeast 129 but the yeast and human TDP1 can also process single strand breaks in vitro 130, 131. The primary substrate for TDP1 is the product of abortive topoisomerase 1 (Top1) reaction, namely, Top1-linked 3′-termini 132, 133. 3′ end processing by TDP1 generates 3′ phosphate which is a substrate for PNK. Finally, under some circumstances some types of blocked 3′ termini could be removed by ERCC1/XPF nuclease, a functional homologue of the yeast Rad1/Rad10 complex proposed to conduct this role in the budding yeast 134.
Aprataxin, a novel DNA end cleaning enzyme, prevents neurodegenerative disease
Aprataxin belongs to a family of nucleotide hydrolases and transferases 135, 136. Mutations in the aprataxin gene causes ataxia oculomotor apraxia, a neurological disorder. Recent evidence has linked aprataxin to DNA repair 137. Aprataxin has been shown to associate with the DNA repair proteins XRCC1 and XRCC4, which are partners of DNA Lig III and Lig IV, respectively 138. Recent studies from Stephen West's laboratory documented a direct role of aprataxin in DNA repair in resolving abortive DNA ligation intermediates 137. Specifically, aprataxin catalyzes the nucleophilic release of adenylate groups covalently linked to 5′ phosphate termini at single strand nicks and gaps, resulting in the production of 5′ phosphate termini that can be efficiently rejoined (Figure 2). The role of aprataxin in DNA repair is supported by the interaction of aprataxin with DNA ligase complexes, and the sensitivity of aprataxin-defective cells to agents that cause DNA single strand breaks, such as ROS.
PALF, a distinct mammalian APE with potentially broad repair function
Akira Yasui's group has recently identified a new protein with APE activity which they have named PALF because of some similarity with aprataxin and PNK 139. PALF accumulates at DNA strand breaks, which is dependent on poly ADP-ribose polymerase (PARP), and directly interacts with the latter. Because PALF interacts with Ku86, XRCC4 and DNA Lig IV, all involved in nonhomologous end joining of DNA double-strand breaks, PALF may have a broad function in repair of both DNA single- and double-strand breaks.
Thus 'end cleaning' is an important intermediate step in both BER and single-strand break repair, to prepare the substrate for subsequent steps, and provides a link between DNA glycosylase and DNA polymerase in BER. The four major DNA end cleaning proteins and their activities are represented in Figure 2.
TREX1 and TREX2, mammalian 3′ exonucleases
TREX1 and TREX2 were identified as DNA 3′ exonucleases ubiquitously expressed in mammalian tissues 140, 141. Their primary function may be in editing during replication by Polβ (and α) which lacks constitutive 3′ exonuclease activity. Recently, TREX1 was indicated to be the product of AGS1 gene whose deficiency causes a severe brain disease 142.
XRCC1 and PARP, single-strand break sensor proteins
XRCC1, a scaffold for recruiting BER proteins
XRCC1 and PARP1 serve as single-strand break sensor proteins. However, XRCC1 functions very differently by acting as a scaffold for recruiting BER proteins for single-strand break repair. XRCC1 physically interacts with several BER proteins and activates many of them. Our results showing interaction of XRCC1 with NEILs implicate XRCC1 in the base excision step of BER as well 51, 115. At the same time, XRCC1 interacts with the end-cleaning enzymes including APE1, PNK and possibly others 126, 143. Moreover, the processing of 5′ OH and/ or 3′ phosphate termini is rate limiting for single strand break repair in XRCC1-mutant cells and cell extracts 126, 135, 144. Its interaction with Polβ and Lig IIIα suggests that XRCC1 is involved primarily in SN-BER 76, 145.
XRCC1 was shown to interact with and to stimulate APE1, and thus may stimulate this activity during end processing 143. However, others did not observe this interaction, and APE1 activity appears to be normal in XRCC1-mutant cells 146, 147.
PARP-1, -2 activated by SSBs
PARP present in mammals but not in E. coli nor yeast transfers ADP-ribose moiety from NAD to a variety of proteins including itself and predominantly histones, nonhistone HMGs and a number of other nuclear proteins including those involved in DNA metabolism to form linear or branched polymer chains using ribose-α (1′-2′) ribose linkages. The branched structure involves ribose-α (1′-1′) ribose linkage. The first ADP-ribose is usually added to the γ-carboxyl residue of Glu in acceptor proteins 148. Some 15 other members of the PARP superfamily including distinct proteins such as tankyrases with functions unrelated to DNA repair have been identified. Genotoxic agents that directly or indirectly generate SSBs in the genome activate PARP-1 and PARP-2 leading to depletion of NAD and indirectly of ATP and induce necrosis. The overlapping function of DNA damage-dependent PARPs, PARP-1 and PARP-2, is essential in mouse cells. PARP1 is recruited very early to the site of DNA strand-break and the activated enzyme may help recruit other repair proteins involved in BER. Poly ADP-ribose (PAR) is removed by the single enzyme, poly(ADP-ribose) glycol hydrolase (PARG) which has both endo- and exoglycosidase activities.
BER in mitochondria
Mitochondrial DNA (mtDNA) contains higher steady-state level of oxidative damage and mutates at rates significantly greater than the nuclear DNA because mitochondria are the predominant site for production of endogenous ROS and tend to accumulate toxic xenobiotics. In addition to their proximity to the sites of ROS generation, it is likely that the mitochondrial genomes are more prone to oxidative damage because histones and other chromatin-associated proteins, present in nuclear genomes and acting as scavengers of oxygen radicals, are absent in the mitochondria.
BER is the only excision repair process active for mitochondrial genomes. All mtDNA repair proteins are nuclear encoded and imported. Most mtDNA repair proteins so far discovered are either identical to nuclear DNA repair proteins or isoforms of nuclear proteins arising from differential splicing 149, 150. The existence of repair of oxidative damage to mtDNA was established several years ago 151, 152, 153. It was also reported that mitochondrial DNA polymerase gamma (Polγ) and mtDNA ligase (Lig IIIβ), which are involved in mtDNA replication are also functional in mitochondrial BER 154, 155. Lig IIIβ is a splice variant of the nuclear Lig IIIα, whereas Polγ is unique for the mitochondria 156. Similarly, splice variants of nuclear OGG1 and NTH1 have been characterized in mitochondria 69, 157. Mitochondrial transport of proteins generally requires an N-terminal mitochondrial targeting sequence (MTS) which is cleaved off in the mitochondrial matrix by specific peptidases 158 although, some mitochondrial proteins lacking MTS, utilize an internal targeting sequence which is not removed during mitochondrial import 159. Further, mitochondrial isozymes are generated primarily to eliminate the nuclear localization signal of such proteins, otherwise required for their nuclear targeting 158.
Although most mitochondrial BER enzymes were characterized and significant BER activity was reported in mitochondrial extracts several years ago 152, 153, 154, the identity of mtAPE, a key BER enzyme, was not established. Recently, our lab showed that the mtAPE lacks the N-terminal 33 amino acid residues including the NLS 150. However, the full-length APE1 could also be detected in mitochondria of some cells. This indicates that deletion of NLS is not absolutely required for mitochondrial import of APE1. However, we have also shown that the extract of mitochondria (containing processing peptidases) but not of nuclei or cytosol, cleaves recombinant hAPE1 to generate an mtAPE-sized product, implying that the removal of NLS is necessary for mitochondrial import of APE. At the same time, deletion mapping of hAPE1 helped us identify a bipartite NLS as well as a nuclear export signal in the N-terminal segment of the APE1 polypeptide 110.
Repair interactome - a new paradigm in BER
The broad view of BER in the last decade was that of a sequential process in which individual repair enzymes carried out reactions independently of one another 112. Based on the emerging evidence for the existence of the 'protein interactome', we propose a new paradigm in BER that postulates collaboration of multiple proteins in coordinated fashion involving specific protein-protein interactions to enhance the efficiency of repair. It is now evident that cellular processes involving repair of both endogenous and induced genomic damage are essential for maintaining genomic integrity and homeostasis, which involve dynamic and complex interactions among a multitude of proteins in the cellular 'proteome' in response to both endogenous cellular activities and environmental stress 160. Such protein complexes are not unique to BER, and similar situations appear to exist for other DNA excision repair pathways. There is increasing evidence that the mammalian BER proteins are organized in temporally controlled complexes for cross-talks among themselves and with DNA, presumably for optimum efficiency of damaged base recognition and repair.
Formation of the multiprotein complexes for oxidative damage repair is enhanced by ROS 51, 161. It was previously shown that NEILs carry out SN-BER mediated by PNK, Pol β, Lig IIIα and XRCC1 by stably interacting with Pol β, Lig IIIα and XRCC1 51, 115. Other interactions such as that between OGG1 and XRCC1 was reported earlier 162. It now appears that additional proteins including those of the DNA replication machinery are also involved in BER 26, 163. Several DNA glycosylases interact with replication related-proteins, namely, PCNA and replication protein A (RPA). MYH and UNG interact with both PCNA and RPA 164, 165, 166. We observed stable and functional association between NEIL1 and the WRN protein 167. Although these associations may be merely for carrying out LP-BER, several observations, e.g., (i) co-localization of the UNG-PCNA-RPA complex at replication foci and (ii) S-phase-specific increase in the levels of UNG2, MYH and NEIL1 50, 165, 168, support our hypothesis that certain glycosylases are involved in RA-BER.
A key unanswered question is whether there is coordinated handover among interacting partners in smaller sub-complexes or a larger complex is preformed to carry out efficient repair. The coordination in protein complex formation was originally compared to the passing of the baton, where the repair product of each enzyme in the BER pathway is “handed” over to the next enzyme in the pathway 169. It would be important to unravel how NEIL1 or any DNA glycosylase can simultaneously and binarily interact with several downstream proteins in the repair pathway initiated by the glycosylase. All of NEIL1's interacting partners identified so far bind to the C-terminus of NEIL1 51, 57, 115. Though NEIL1's interaction with its multiple partners using a common interaction interface (C-terminus) may be explained based on the unfolded conformation / flexibility of its C-terminus, further structural characterization of the C-terminus of NEIL1 and structure-function studies are required to elucidate the mechanism in detail.
Unique Structural features of mammalian DNA glycosylases
Mammalian DNA glycosylases possess unique structural features compared with their counterparts in lower organisms and bacteria. The major characteristic of mammalian glycosylases (which may also be true for other mammalian BER enzymes) is an unfolded extension or tail which participates in subcellular translocation, and more importantly in protein-protein interactions. The sequence alignment of hNTH1 with NTHs from E. coli, archea and other lower organisms shows that the human (and other mammalian) NTH1 has an N-terminal tail, absent in E. coli and archea. This N-terminal tail (residues 1-95) contains putative nuclear and mitochondrial localization signals 170. hNTH1 was found to have low activity compared to E. coli Nth 49, 171. This is because of the inhibitory role of N-terminal tail of hNTH1 on its turnover. Thus, the N-terminal tail of hNTH1 could regulate the overall catalytic turnover by reducing the rate of product release without affecting glycosylase or AP lyase activities 172.
The crystal structure of an enzymatically active deletion construct of hNEIL1 (lacking 56 C-terminal residues) and other modeling studies indicated that hNEIL1 has an unfolded C-terminal extension (∼100 residues) which is absent in bacterial Nei 173. The crystal structure of hNEIL1 also indicates the presence of internal Zn-less finger, unlike the C-terminal Zn finger present in other Nei orthologs. Our data show that NEIL1 interacts with downstream SN-BER proteins, via its unfolded C-terminal extension (residues 289-349) 115.
We used the software of Molecular Kinetics, Inc. 174 for prediction of naturally disordered regions (PONDR) to compare the secondary structures of hNEIL1 and hNTH1 with their E. coli prototypes Nei and Nth respectively (Figure 3). Extended disordered regions are present only in the human proteins and their locations at the N-terminus of hNTH1 and at the C-terminus of hNEIL1 are consistent with the experimental evidence showing the absence of defined structure in these regions using X-ray crystallography. The prediction about the disordered structure in the C-terminal region of NEIL1 supports the hypothesis that the region individually interacting with multiple proteins is intrinsically disordered.
PONDR plot of the predicted secondary structures of hNEIL1, hNTH1 and their E. coli prototypes, endonuclease VIII (Nei) and endonuclease III (Nth) respectively. The protein sequences were obtained from NCBI database. PONDR score of 0.5 and higher indicates disordered structure 174. The disordered C-terminal segment of hNEIL1 and N-terminal segment of hNTH1 are depicted by wiggly lines.
Figure 3 shows the PONDR©plot of the predicted secondary structures in hNEIL1 and hNTH1. The disordered structures of hNEIL1 at the C-terminus and of hNTH1 at the N-terminus are absent in their E. coli prototypes.
DNA glycosylases interact with non-BER proteins
Several other non-BER proteins have also been shown to be involved in BER, however their precise in vivo role in repair is yet to be understood. NEIL2 interacts with YB-1, a Y-box binding protein, and it was suggested that YB-1 may be required for the fine-tuning of repair 161. NTH1 was also shown to interact with and be stimulated by YB-1 175. We have recently shown a stable interaction between NEIL1 and the 9-1-1 complex and the stimulation of NEIL1 as a result of this interaction 75. 9-1-1 is a heterotrimeric protein complex and it acts as a sliding clamp similar to PCNA, but is activated by DNA damage and cell cycle checkpoints. NEIL1's interaction with 9-1-1 highlights the linkage between NEIL1-initiated BER and damage signaling pathways. Our recent studies also showed NEIL1's stable association with PCNA 74. It is highly likely that NEIL1's interactome contains a large number of non-BER proteins. Whether and how these proteins participate in BER remains an important topic for future investigations.
Covalent modifications of DNA glycosylases and modulation of BER
Regulation of BER may be further fine-tuned through posttranslational modifications of the BER proteins. Such modifications include phosphorylation, acetylation, mono- or poly-ADP-ribosylation, mono- and poly-ubiquitylation, sumoylation and methylation among others 176. Covalent modifications could have multiple physiological effects on proteins, including modulation of stability, interaction, organelle targeting, and enzymatic activity etc. 177. Such studies are still in an early stage. Some initial observations on modifications of mammalian DNA glycosylases are summarized as follows.
Sumoylation of TDG
Sumoylation and ubiquitylation alter activities of target proteins 178. SUMO-1 and -3 modify TDG, a glycosylase that removes uracil (or thymine) from its substrate U(T)•G pair 179. TDG remains bound to the AP site after the excision of uracil and sumoylation reduces its affinity, thus releasing the product. The modified TDG is recycled by deconjugation, restoring its activity.
Acetylation of DNA glycosylases
The presence of multiple species of OGG1 in 2-D gel analysis of HeLa extracts suggested its covalent modification, which we showed to be acetylation 180. We also showed that about 20% of OGG1 is acetylated in HeLa cells, and identified two acetylation acceptor lysine residues (Lys 338 and Lys 341). We further showed that acetylation enhances OGG1 turnover by decreasing its affinity for the product. We earlier observed that APE1 enhances OGG1 turnover which is higher when OGG1 is acetylated 39. ROS induced an increase in the AcOGG1 level, indirectly through activation of p300. We proposed that acetylation of OGG1 provides a mechanism for rapid cellular response to handle the increased load of oxidized bases induced by ROS 180.
Studies from our lab showed that NEIL2 is also acetylated, predominantly at Lys 49 and 153 181. Lys 49 is conserved among Fpg family members and its mutation inactivates the enzyme. Lys 49 acetylation strongly inhibits NEIL2 activity. We have proposed that acetylation at Lys 49 acts as a regulatory switch for NEIL2's activity, and Lys 153 acetylation could regulate its interactions.
It has been shown that TDG is acetylated both in vivo and in vitro by CBP/p300 which forms a functional complex with TDG 182. Strong acetylation sites were identified at Lys residues 70, 94, 95 and 98. TDG acetylation does not affect binding and cleavage of G•T, G•U mispaired DNA but leads to release of CBP/p300 from the DNA bound complex and abrogates the interaction with APE suggesting a role in suppression of APE-dependent repair by blocking the next reaction that normally follows the removal of the mispaired base. Thus acetylation appears to deregulate TDG-coupled DNA repair and contribute to the genomic instability commonly associated with cancer.
Summary and future perspectives
BER is distinct from the other excision repair pathways because the first step of lesion recognition by a DNA glycosylase is unique. This review on the early steps in base excision and strand break repair aims at highlighting the key roles of DNA glycosylases and APE1 (and other end-cleaning activities) in maintenance of genomic integrity in mammalian genomes. Furthermore, BER represents the predominant process for repairing endogenous DNA lesions mostly induced by ROS. While BER was earlier believed to be the simplest repair process, its complexities are now becoming increasingly evident. In this review, we have touched upon a few of these complexities including interaction of BER enzymes with non-BER proteins, their covalent modifications, formation of pair-wise and multiprotein repair complexes and its regulation. Future research will undoubtedly be focused on these and other aspects of BER. For example, we have shown ROS-mediated activation of NEIL1 and APE1 183, 184. We have shown that ROS enhances NEIL2 complex formation 161. ROS also enhances modification of OGG1 180 and possibly other BER proteins. How such modifications affect repair efficiency in vivo should be addressed. Preliminary studies showing preferential repair of oxidized bases in replicating genomes need to be extended. The linkage of BER to susceptibility to various diseases also warrants extensive exploration. Finally, the mechanism of recognition of BER substrates, which cause subtle deformations in DNA structure in chromatin, followed by their repair requiring access of the repair complex to the lesion needs to be explored. These challenges will keep the investigators busy for many years.
Abbreviations
- (AP):
-
Abbreviations: abasic
- (APE):
-
AP-endonuclease
- (BER):
-
base excision repair
- (dRP):
-
deoxyribose phosphate
- (8-oxoG):
-
8-oxo-7,8 dihydroguanine
- (5-OHU):
-
5-hydroxyuracil
- (Fpg):
-
formamidopyrimidine-DNA glycosylase
- (Lig I):
-
DNA ligase I
- (Lig IIIα):
-
DNA ligase IIIα
- (LP-BER):
-
long-patch BER
- (MGMT):
-
O6-methylguanine-DNA methyltransferase
- (MPG):
-
N-methylpurine-DNA glycosylase
- (mtDNA):
-
mitochondrial DNA
- (MTS):
-
mitochondrial targeting sequence
- (Nfo):
-
endonuclease IV
- (Nei):
-
endonuclease VIII
- (NLS):
-
nuclear localization signal
- (Nth):
-
endonuclease III
- (O2):
-
superoxide radical anion
- (OGG1):
-
8-oxoguanine-DNA glycosylase
- (OH•):
-
hydroxy radical
- (PARG):
-
poly(ADP-ribose) glycol hydrolase
- (PARP):
-
poly(ADP-ribose) polymerase
- (PNK):
-
polynucleotide kinase
- (Polγ):
-
mitochondrial DNA polymerase gamma
- (Polβ):
-
DNA polymerase β
- (PONDR):
-
prediction of naturally disordered regions
- (RA-BER):
-
replication-associated BER
- (RFC):
-
replication factor-C
- (ROS):
-
reactive oxygen species
- (RPA):
-
replication protein A
- (SN-BER):
-
single nucleotide BER
- (SNP):
-
single nucleotide polymorphisms
- (TDG):
-
thymine-(T•G)-DNA glycosylase
- (TLS):
-
translesion synthesis
- (Top1):
-
topoisomerase 1
- (3′ PUA):
-
3′ phospho α,β-unsaturated aldehyde [4-hydroxylpentenal]
- (TDP1):
-
Tyrosyl phosphodiesterase 1
- (UDG):
-
uracil DNA glycosylase
- (Xth):
-
exonuclease III
References
Dawson TL, Gores GJ, Nieminen AL, Herman B, Lemasters JJ . Mitochondria as a source of reactive oxygen species during reductive stress in rat hepatocytes. Am J Physiol 1993; 264:C961–C967.
Griendling KK, Sorescu D, Ushio-Fukai M . NAD(P)H oxidase: role in cardiovascular biology and disease. Circ Res 2000; 86:494–501.
Lindahl T . Instability and decay of the primary structure of DNA. Nature 1993; 362:709–715.
Prakash S, Johnson RE, Prakash L . Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem 2005; 74:317–353.
Wallace DJ . A reason for the rarity of male lupus. Lupus 1998; 7:60.
Levine AJ . p53, the cellular gatekeeper for growth and division. Cell 1997; 88:323–331.
Helbock HJ, Beckman KB, Ames BN . 8-Hydroxydeoxyguanosine and 8-hydroxyguanine as biomarkers of oxidative DNA damage. Methods Enzymol 1999; 300:156–166.
Shibutani S, Takeshita M, Grollman AP . Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature 1991; 349:431–434.
Michaels ML, Miller JH . The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J Bacteriol 1992; 174:6321–6325.
Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T . DNA Repair and Mutagenesis. 2nd Edition. Washington: ASM Press, 2006.
Krokan HE, Nilsen H, Skorpen F, Otterlei M, Slupphaug G . Base excision repair of DNA in mammalian cells. FEBS Lett 2000; 476:73–77.
Papadopoulos N, Lindblom A . Molecular basis of HNPCC: mutations of MMR genes. Hum Mutat 1997; 10:89–99.
Breen AP, Murphy JA . Reactions of oxyl radicals with DNA. Free Rad Biol Med 1995; 18:1033–1077.
Demple B, DeMott MS . Dynamics and diversions in base excision DNA repair of oxidized abasic lesions. Oncogene 2002; 21:8926–8934.
Mitra S, Hazra TK, Roy R, et al. Complexities of DNA base excision repair in mammalian cells. Mol cells 1997; 7:305–312.
Doetsch PW, Cunningham RP . The enzymology of apurinic/apyrimidinic endonucleases. Mutat Res 1990; 236:173–201.
Matsumoto Y, Kim K . Excision of deoxyribose phosphate residues by DNA polymerase beta during DNA repair. Science 1995; 269:699–702.
Prasad R, Beard WA, Strauss PR, Wilson SH . Human DNA polymerase beta deoxyribose phosphate lyase. Substrate specificity and catalytic mechanism. J Biol Chem 1998; 273:15263–15270.
Lindahl T . An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc Natl Acad Sci USA 1974; 71:3649–3653.
Krokan HE, Drablos F, Slupphaug G . Uracil in DNA-occurrence, consequences and repair. Oncogene 2002; 21:8935–8948.
Slupphaug G, Mol CD, Kavli B, et al. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature 1996; 384:87–92.
Parker JB, Bianchet MA, Krosky DJ, et al. Enzymatic capture of an extrahelical thymine in the search for uracil in DNA. Nature 2007; 449:433–437.
Masaoka A, Matsubara M, Hasegawa R, et al. Mammalian 5-formyluracil-DNA glycosylase. 2. Role of SMUG1 uracil-DNA glycosylase in repair of 5-formyluracil and other oxidized and deaminated base lesions. Biochemistry 2003; 42:5003–5012.
Wibley JE, Waters TR, Haushalter K, Verdine GL, Pearl LH . Structure and specificity of the vertebrate anti-mutator uracil-DNA glycosylase SMUG1. Mol Cell 2003; 11:1647–1659.
Hardeland U, Bentle M, Jiricny J, Schar P . The versatile thymine DNA-glycosylase: a comparative characterization of the human, Drosophila and fission yeast orthologs. Nucleic Acids Res 2003; 31:2261–2271.
Mitra S, Boldogh I, Izumi T, Hazra TK . Complexities of the DNA base excision repair pathway for repair of oxidative DNA damage. Environ Mol Mutagen 2001; 38:180–190.
Mitra S . MGMT: a personal perspective. DNA Repair (Amst) 2007; 6:1064–1070.
Mitra S . In: Kaina B, Margison GP, eds. MGMT: Reversal of fortune - from basic biochemistry to clinical exploitation of the DNA damage reversal protein, O6-methylguanine-DNA methyltransferase. DNA Repair (Amst) 2007; 6:1063–1228.
Samson L, Cairns J . A new pathway for DNA repair in Escherichia coli. Nature 1977; 267:281–283.
Sedgwick B, Bates PA, Paik J, Jacobs SC, Lindahl T . Repair of alkylated DNA: recent advances. DNA Repair (Amst) 2007; 6:429–442.
Mitra S, Kaina B . Regulation of repair of alkylation damage in mammalian genomes. Prog Nucleic Acids Res Mol Biol 1993; 44:109–142.
Roy R, Kennel SJ, Mitra S . Distinct substrate preference of human and mouse N-methylpurine-DNA glycosylases. Carcinogenesis 1996; 17:2177–2182.
Lu AL, Chang DY . Repair of single base-pair transversion mismatches of Escherichia coli in vitro: correction of certain A/G mismatches is independent of dam methylation and host mutHLS gene functions. Genetics 1988; 118:593–600.
Nghiem Y, Cabrera M, Cupples CG, Miller JH . The mutY gene: a mutator locus in Escherichia coli that generates G.C-T.A transversions. Proc Natl Acad Sci USA 1988; 85:2709–2713.
McGoldrick JP, Yeh YC, Solomon M, Essignmann JM, Lu AL . Characterization of a mammalian homolog of the E. coli MutY mismatch repair protein. Mol Cell Biol 1995; 15:989–996.
Ohtsubo T, Nishioka K, Imaiso Y, et al. Identification of human MutY homolog (hMYH) as a repair enzyme for 2-hydroxyadenine in DNA and detection of multiple forms of hMYH located in nuclei and mitochondria. Nucleic Acids Res 2000; 15:1355–1364.
McCullough AK, Dodson ML, Lloyd RS . Initiation of base excision repair: glycosylase mechanisms and structures. Annu Rev Biochem 1999; 68:255–285.
Radicella JP, Dherin C, Desmaze C, Fox MS, Boiteux S . Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci USA 1997; 94:8010–8015.
Hill JW, Hazra TK, Izumi T, Mitra S . Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease: potential coordination of the initial steps in base excision repair. Nucleic Acids Res 2001; 29:430–438.
Huffman JL, Sundheim O, Tainer JA . DNA base damage recognition and removal: new twists and grooves. Mutat Res 2005; 577:55–76.
Zharkov DO, Shoham G, Grollman AP . Structural characterization of the Fpg family of DNA glycosylases. DNA Repair (Amst) 2003; 2:839–862.
Guan Y, Manuel RC, Arvai AS, et al. MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nat Struct Biol 1998; 5:1058–1064.
Bruner SD, Norman DP, Verdine GL . Structural basis recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature 2000; 403:859–866.
Hitomi K, Iwai S, Tainer JA . The intricate structural chemistry of base excision repair machinery: implications for DNA damage recognition, removal, and repair. DNA Repair (Amst) 2007; 6:410–428.
Sugahara M, Mikawa T, Kumasaka T, et al. Crystal structure of a repair enzyme of oxidatively damaged DNA, MutM (Fpg), from an extreme thermophile, Thermus thermophilus HB8. EMBO J 2000; 19:3857–3869.
Gilboa R, Zharkov DO, Golan G, et al. Structure of formamidopyrimidine-DNA glycosylase covalently complexed to DNA. J Biol Chem 2002; 277:19811–19816.
Zharkov DO, Golan G, Gilboa R, et al. Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. EMBO J 2002; 21:789–800.
Aburatani H, Hippo Y, Ishida T, et al. Cloning and characterization of mammalian 8-hydroxyguanine-specific DNA glycosylase/apurinic, apyrimidinic lyase, a functional mutM homologue. Cancer Res 1997; 57:2151–2156.
Ikeda S, Biswas T, Roy R, et al. Purification and characterization of human NTH1, a homolog of Escherichia coli endonuclease III. Direct identification of Lys-212 as the active nucleophilic residue. J Biol Chem 1998; 273:21585–21593.
Hazra TK, Izumi T, Boldogh I, et al. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc Natl Acad Sci USA 2002; 99:3523–3528.
Das A, Wiederhold L, Leppard JB, et al. NEIL2-initiated, APE-independent repair of oxidized bases in DNA: Evidence for a repair complex in human cells. DNA Repair (Amst) 2006; 5:1439–1448.
Bandaru V, Sunkara S, Wallace SS, Bond JP . A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair (Amst) 2002; 1:517–529.
Hazra TK, Kow YW, Hatahet Z, et al. Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. J Biol Chem 2002; 277:30417–30420.
Morland I, Rolseth V, Luna L, et al. Human DNA glycosylases of the bacterial Fpg/MutM superfamily: an alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res 2002; 30:4926–4936.
Takao M, Kanno S, Kobayashi K, et al. A back-up glycosylase in Nth1 knock-out mice is a functional Nei (endonuclease VIII) homologue. J Biol Chem 2002; 277:42205–42213.
Parsons JL, Zharkov DO, Dianov GL . NEIL1 excises 3′ end proximal oxidative DNA lesions resistant to cleavage by NTH1 and OGG1. Nucleic Acids Res 2005; 33:4849–4856.
Dou H, Mitra S, Hazra TK . Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J Biol Chem 2003; 278:49679–49684.
Alseth I, Eide L, Pirovano M, et al. The Saccharomyces cerevisiae homologues of endonuclease III from Escherichia coli, Ntg1 and Ntg2, are both required for efficient repair of spontaneous and induced oxidative DNA damage in yeast. Mol Cell Biol 1999; 19:3779–3787.
Klungland A, Rosewell I, Hollenbach S, et al. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc Natl Acad Sci USA 1999; 96:13300–13305.
Minowa O, Arai T, Hirano M, et al. Mmh/Ogg1 gene inactivation results in accumulation of 8-hydroxyguanine in mice. Proc Natl Acad Sci USA 2000; 97:4156–4161.
Osterod M, Hollenbach S, Hengstler JG, et al. Age-related and tissue-specific accumulation of oxidative DNA base damage in 7,8-dihydro-8-oxoguanine-DNA glycosylase (Ogg1) deficient mice. Carcinogenesis 2001; 22:1459–1463.
Arai T, Kelly VP, Minowa O, Noda T, Nishimura S . High accumulation of oxidative DNA damage, 8-hydroxyguanine, in Mmh/Ogg1 deficient mice by chronic oxidative stress. Carcinogenesis 2002; 23:2005–2010.
Gu H, Marth JD, Orban PC, Mossmann H, Rajewsky K . Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science 1994; 265:103–106.
Xanthoudakis S, Smeyne RJ, Wallace JD, Curran T . The redox/DNA repair protein, Ref-1, is essential for early embryonic development in mice. Proc Natl Acad Sci USA 1996; 93:8919–8923.
Modrich P, Lahue R . Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem 1996; 65:101–133.
Friedberg EC, Aguilera A, Gellert M, et al. DNA repair: from molecular mechanism to human disease. DNA Repair (Amst) 2006; 5:986–996.
Crew KD, Gammon MD, Terry MB, et al. Polymorphisms in nucleotide excision repair genes, polycyclic aromatic hydrocarbon-DNA adducts, and breast cancer risk. Cancer Epidemiol Biomarkers Prev 2007; 16:2033–2041.
Mitra S, Izumi T, Boldogh I, et al. Choreography of oxidative damage repair in mammalian genomes. Free Radic Biol Med 2002; 33:15–28.
Karahalil B, de Souza-Pinto NC, Parsons JL, Elder RH, Bohr VA . Compromised incision of oxidized pyrimidines in liver mitochondria of mice deficient in NTH1 and OGG1 glycosylases. J Biol Chem 2003; 278:33701–33707.
Reardon JT, Bessho T, Kung HC, Bolton PH, Sancar A . In vitro repair of oxidative DNA damage by human nucleotide excision repair system: possible explanation for neurodegeneration in xeroderma pigmentosum patients. Proc Natl Acad Sci USA 1997; 94:9463–9468.
Hazra TK, Das A, Das S, et al. Oxidative DNA damage repair in mammalian cells: a new perspective. DNA Repair (Amst) 2007; 6:470–480.
Haug T, Skorpen F, Aas PA, et al. Regulation of expression of nuclear and mitochondrial forms of human uracil-DNA glycosylase. Nucleic Acids Res 1998; 26:1449–1457.
Kavli B, Sundheim O, Akbari M, et al. hUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. J Biol Chem 2002; 277:39926–39936.
Dou H, Theriot CA, Das A, et al. Interaction of the human DNA glycosylase NEIL1 with proliferating cell nuclear antigen: the potential for replication-associated repair of oxidized bases in mammalian genomes. J Biol Chem 2007 Nov 21; doi:10.1074/jbc.M709186200.
Guan X, Bai H, Shi G, et al. The human checkpoint sensor Rad9-Rad1-Hus1 interacts with and stimulates NEIL1 glycosylase. Nucleic Acids Res 2007; 35:2463–2472.
Kubota Y, Nash RA, Klungland A, et al. Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. EMBO J 1996; 15:6662–6670.
Frosina G, Fortini P, Rossi O, et al. Two pathways for base excision repair in mammalian cells. J Biol Chem 1996; 271:9573–9578.
Matsumoto T, Yokote K, Tamura K, et al. Platelet-derived growth factor activates p38 mitogen-activated protein kinase through a Ras-dependent pathway that is important for actin reorganization and cell migration. J Biol Chem 1999; 274:13954–13960.
Matsumoto Y . Molecular mechanism of PCNA-dependent base excision repair. Prog Nucleic Acid Res Mol Biol 2001; 68:129–138.
Klungland A, Lindahl T . Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN-1). EMBO J 1997; 16:3341–3348.
Piersen CE, Prasad R, Wilson SH, Lloyd RS . Evidence for an imino intermediate in the DNA polymerase beta deoxyribose phosphate excision reaction. J Biol Chem 1996; 271:17811–17815.
Levin DS, Vijayakumar S, Liu X, et al. A conserved interaction between the replicative clamp loader and DNA ligase in eukaryotes: implications for Okazaki fragment joining. J Biol Chem 2004; 279:55196–55201.
Singh P, Zheng L, Chavez V, Qiu J, Shen B . Concerted action of exonuclease and Gap-dependent endonuclease activities of FEN-1 contributes to the resolution of triplet repeat sequences (CTG)n- and (GAA)n-derived secondary structures formed during maturation of Okazaki fragments. J Biol Chem 2007; 282:3465–3477.
Levin DS, McKenna AE, Motycka TA, Matsumoto Y, Tomkinson AE . Interaction between PCNA and DNA ligase I is critical for joining of Okazaki fragments and long-patch base-excision repair. Curr Biol 2000; 10:919–922.
Ochs K, Sobol RW, Wilson SH, Kaina B . Cells deficient in DNA polymerase beta are hypersensitive to alkylating agent-induced apoptosis and chromosomal breakage. Cancer Res 1999; 59:1544–1551.
Liu Y, Kao HI, Bambara RA . Flap endonuclease 1: a central component of DNA metabolism. Annu Rev Biochem 2004; 73:589–615.
Haring M, Rudiger H, Demple B, Boiteux S, Epe B . Recognition of oxidized abasic sites by repair endonucleases. Nucleic Acids Res 1994; 22:2010–2015.
Hanna M, Chow BL, Morey NJ, Jinks-Robertson S, Doetsch PW, Xiao W . Involvement of two endonuclease III homologs in the base excision repair pathway for the processing of DNA alkylation damage in Saccharomyces cerevisiae. DNA Repair (Amst) 2004; 3:51–59.
Demple B, Harrison L . Repair of oxidative damage to DNA: enzymology and biology. Annu Rev Biochem 1994; 63:915–948.
Demple B, Johnson A, Fung D . Exonuclease III and endonuclease IV remove 3′ blocks from DNA synthesis primers in H2O2-damaged Escherichia coli. Proc Natl Acad Sci USA 1986; 83:7731–7735.
Ljungquist S . A new endonuclease from Escherichia coli acting at apurinic sites in DNA. J Biol Chem 1977; 252:2808–2814.
Cunningham RP, Saporito SM, Spitzer SG, Weiss B . Endonuclease IV (nfo) mutant of Escherichia coli. J Bacteriol 1986; 168:1120–1127.
Auerbach P, Bennett RA, Bailey EA, Krokan HE, Demple B . Mutagenic specificity of endogenously generated abasic sites in Saccharomyces cerevisiae chromosomal DNA. Proc Natl Acad Sci USA 2005; 102:17711–17716.
Zhang Y, Yuan F, Wu X, et al. Error-free and error-prone lesion bypass by human DNA polymerase kappa in vitro. Nucleic Acids Res 2000; 28:4138–4146.
Kokoska RJ, McCulloch SD, Kunkel TA . The efficiency and specificity of apurinic/apyrimidinic site bypass by human DNA polymerase eta and Sulfolobus solfataricus Dpo4. J Biol Chem 2003; 278:50537–50545.
Smith CE, Llorente B, Symington LS . Template switching during break-induced replication. Nature 2007; 447:102–105.
Selby CP, Sancar A . Structure and function of the (A)BC excinuclease of Escherichia coli. Mutat Res 1990; 236:203–211.
Popoff SC, Spira AI, Johnson AW, Demple B . Yeast structural gene (APN1) for the major apurinic endonuclease: homology to Escherichia coli endonuclease IV. Proc Natl Acad Sci USA 1990; 87:4193–4197.
Johnson RE, Torres-Ramos CA, Izumi T et al. Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev 1998; 12:3137–3143.
Guillet M, Boiteux S . Endogenous DNA abasic sites cause cell death in the absence of Apn1, Apn2 and Rad1/Rad10 in Saccharomyces cerevisiae. EMBO J 2002; 21:2833–2841.
Gorman MA, Morera S, Rothwell DG, et al. The crystal structure of the human DNA repair endonuclease HAP1 suggests the recognition of extra-helical deoxyribose at DNA abasic sites. EMBO J 1997; 16:6548–6558.
Mol CD, Izumi T, Mitra S, Tainer JA . DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination. Nature 2000; 403:451–456.
Oezguen N, Schein CH, Peddi SR, et al. A „moving metal mechanism“ for substrate cleavage by the DNA repair endonuclease APE-1. Proteins 2007; 68:313–323.
Ribar B, Izumi T, Mitra S . The major role of human AP-endonuclease homolog Apn2 in repair of abasic sites in Schizosaccharomyces pombe. Nucleic Acids Res 2004; 32:115–126.
Ivanov I, Tainer JA, McCammon JA . Unraveling the three-metal-ion catalytic mechanism of the DNA repair enzyme endonuclease IV. Proc Natl Acad Sci USA 2007; 104:1465–1470.
Demple B, Herman T, Chen DS . Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. Proc Natl Acad Sci USA 1991; 88:11450–11454.
Hadi MZ, Wilson DM, 3rd. Second human protein with homology to the Escherichia coli abasic endonuclease exonuclease III. Environ Mol Mutagen 2000; 36:312–324.
Tsuchimoto D, Sakai Y, Sakumi K, et al. Human APE2 protein is mostly localized in the nuclei and to some extent in the mitochondria, while nuclear APE2 is partly associated with proliferating cell nuclear antigen. Nucleic Acids Res 2001; 29:2349–2360.
Izumi T, Mitra S . Deletion analysis of human AP-endonuclease: minimum sequence required for the endonuclease activity. Carcinogenesis 1998; 19:525–527.
Jackson EB, Theriot CA, Chattopadhyay R, Mitra S, Izumi T . Analysis of nuclear transport signals in the human apurinic/apyrimidinic endonuclease (APE1/Ref1). Nucleic Acids Res 2005; 33:3303–3312.
Kelley MR, Parsons SH . Redox regulation of the DNA repair function of the human AP endonuclease Ape1/ref-1. Antioxid Redox Signal 2001; 3:671–683.
Izumi T, Wiederhold LR, Roy G, et al. Mammalian DNA base excision repair proteins: their interactions and role in repair of oxidative DNA damage. Toxicology 2003; 193:43–65.
Bhakat KK, Izumi T, Yang SH, Hazra TK, Mitra S . Role of acetylated human AP-endonuclease (APE1/Ref-1) in regulation of the parathyroid hormone gene. EMBO J 2003; 22:6299–6309.
Marenstein DR, Wilson DM, 3rd, Teebor GW. Human AP endonuclease (APE1) demonstrates endonucleolytic activity against AP sites in single-stranded DNA. DNA Repair (Amst) 2004; 3:527–533.
Wiederhold L, Leppard JB, Kedar P, et al. AP endonuclease-independent DNA base excision repair in human cells. Mol Cell 2004; 15:209–220.
Wong D, DeMott MS, Demple B . Modulation of the 3′→5′-exonuclease activity of human apurinic endonuclease (Ape1) by its 5′-incised Abasic DNA product. J Biol Chem 2003; 278:36242–36249.
Meira LB, Devaraj S, Kisby GE, et al. Heterozygosity for the mouse Apex gene results in phenotypes associated with oxidative stress. Cancer Res 2001; 61:5552–5557.
Sugo N, Aratani Y, Nagashima Y, Kubota Y, Koyama H . Neonatal lethality with abnormal neurogenesis in mice deficient in DNA polymerase beta. EMBO J 2000; 19:1397–1404.
Sobol RW, Horton JK, Kuhn R, et al. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature 1996; 379:183–186.
Izumi T, Brown DB, Naidu CV, et al. Two essential but distinct functions of the mammalian abasic endonuclease. Proc Natl Acad Sci USA 2005; 102:5739–5743.
Fung H, Demple B . A vital role for Ape1/Ref1 protein in repairing spontaneous DNA damage in human cells. Mol Cell 2005; 17:463–470.
Evans AR, Limp-Foster M, Kelley MR . Going APE over ref-1. Mutat Res 2000; 461:83–108.
Izumi T, Hazra TK, Boldogh I, et al. Requirement for human AP endonuclease 1 for repair of 3′-blocking damage at DNA single-strand breaks induced by reactive oxygen species. Carcinogenesis 2000; 21:1329–1334.
Mitra S, Izumi T, Boldogh I, et al. Intracellular trafficking and regulation of mammalian AP-endonuclease 1 (APE1), an essential DNA repair protein. DNA Repair (Amst) 2007; 6:461–469.
Meijer M, Karimi-Busheri F, Huang TY, Weinfeld M, Young D . Pnk1, a DNA kinase/phosphatase required for normal response to DNA damage by gamma-radiation or camptothecin in Schizosaccharomyces pombe. J Biol Chem 2002; 277:4050–4055.
Whitehouse CJ, Taylor RM, Thistlethwaite A, et al. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell 2001; 104:107–117.
Mokkapati SK, Wiederhold L, Hazra TK, Mitra S . Stimulation of DNA glycosylase activity of OGG1 by NEIL1: functional collaboration between two human DNA glycosylases. Biochemistry 2004; 43:11596–11604.
Caldecott KW . Mammalian single-strand break repair: mechanisms and links with chromatin. DNA Repair (Amst) 2007; 6:443–453.
Vance JR, Wilson TE . Yeast Tdp1 and Rad1-Rad10 function as redundant pathways for repairing Top1 replicative damage. Proc Natl Acad Sci USA 2002; 99:13669–13674.
Pouliot JJ, Robertson CA, Nash HA . Pathways for repair of topoisomerase I covalent complexes in Saccharomyces cerevisiae. Genes Cells 2001; 6:677–687.
El-Khamisy SF, Saifi GM, Weinfeld M, et al. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature 2005; 434:108–113.
Yang SW, Burgin AB, Jr ., Huizenga BN, et al. A eukaryotic enzyme that can disjoin dead-end covalent complexes between DNA and type I topoisomerases. Proc Natl Acad Sci USA 1996; 93:11534–11539.
Pouliot JJ, Yao KC, Robertson CA, Nash HA . Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I complexes. Science 1999; 286:552–555.
Guzder SN, Torres-Ramos C, Johnson RE, et al. Requirement of yeast Rad1-Rad10 nuclease for the removal of 3′-blocked termini from DNA strand breaks induced by reactive oxygen species. Genes Dev 2004; 18:2283–2291.
Ahel I, Rass U, EI-Khamisy SF, et al. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature 2006; 443:713–716.
Brenner C . Hint, Fhit, and GalT: function, structure, evolution, and mechanism of three branches of the histidine triad superfamily of nucleotide hydrolases and transferases. Biochemistry 2002; 41:9003–9014.
Rass U, Ahel I, West SC . Actions of aprataxin in multiple DNA repair pathways. J Biol Chem 2007; 282:9469–9474.
Sano Y, Date H, Igarashi S, et al. Aprataxin, the causative protein for EAOH is a nuclear protein with a potential role as a DNA repair protein. Ann Neurol 2004; 55:241–249.
Kanno S, Kuzuoka H, Sasao S, et al. A novel human AP endonuclease with conserved zinc-finger-like motifs involved in DNA strand break responses. EMBO J 2007; 26:2094–2103.
Mazur DJ, Perrino FW . Structure and expression of the TREX1 and TREX2 3′ → 5′ exonuclease genes. J Biol Chem 2001; 276:14718–14727.
Hoss M, Robins P, Naven TJ, et al. A human DNA editing enzyme homologous to the Escherichia coli DnaQ/MutD protein. EMBO J 1999; 18:3868–3875.
Crow YJ, Hayward BE, Parmar R, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat Genet 2006; 38:917–920.
Vidal AE, Boiteux S, Hickson ID, Radicella JP . XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interactions. EMBO J 2001; 20:6530–6539.
Lan L, Nakajima S, Oohata Y, et al. In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc Natl Acad Sci USA 2004; 101:13738–13743.
Caldecott KW, McKeown CK, Tucker JD, Ljungquist S, Thompson LH . An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol Cell Biol 1994; 14:68–76.
Fan J, Otterlei M, Wong HK, Tomkinson AE, Wilson DM 3rd. XRCC1 co-localizes and physically interacts with PCNA. Nucleic Acids Res 2004; 32:2193–2201.
Wong HK, Kim D, Hogue BA, McNeill DR, Wilson DM 3rd. DNA damage levels and biochemical repair capacities associated with XRCC1 deficiency. Biochemistry 2005; 44:14335–14343.
Schreiber V, Dantzer F, Ame JC, de Murcia G . Poly(ADP-ribose): novel functions for an old molecule. Nat Rev 2006; 7:517–528.
Stuart JA, Hashiguchi K, Wilson DM, et al. DNA base excision repair activities and pathway function in mitochondrial and cellular lysates from cells lacking mitochondrial DNA. Nucleic Acids Res 2004; 32:2181–2192.
Chattopadhyay R, Wiederhold L, Szczesny B, et al. Identification and characterization of mitochondrial abasic (AP)-endonuclease in mammalian cells. Nucleic Acids Res 2006; 34:2067–2076.
Driggers WJ, LeDoux SP, Wilson GL . Repair of oxidative damage within the mitochondrial DNA of RINr 38 cells. J Biol Chem 1993; 268:22042–22045.
Dianov GL, Souza-Pinto N, Nyaga SG, et al. Base excision repair in nuclear and mitochondrial DNA. Prog Nucleic Acids Res Mol Biol 2001; 68:285–297.
LeDoux SP, Wilson GL . Base excision repair of mitochondrial DNA damage in mammalian cells. Prog Nucleic Acid Res Mol Biol 2001; 68:273–284.
Pinz KG, Bogenhagen DF . Efficient repair of abasic sites in DNA by mitochondrial enzymes. Mol Cell Biol 1998; 18:1257–1265.
Kaguni LS . DNA polymerase gamma, the mitochondrial replicase. Annu Rev Biochem 2004; 73:293–320.
Tomkinson AE, Mackey ZB . Structure and function of mammalian DNA ligases. Mutat Res 1998; 407:1–9.
Nishioka K, Ohtsubo T, Oda H, et al. Expression and differential intracellular localization of two major forms of human 8-oxoguanine DNA glycosylase encoded by alternatively spliced OGG1 mRNAs. Mol Biol Cell 1999; 10:1637–1652.
Pfanner N . Protein sorting: recognizing mitochondrial presequences. Curr Biol 2000; 10:R412–415.
Diekert K, Kispal G, Guiard B, Lill R . An internal targeting signal directing proteins into the mitochondrial intermembrane space. Proc Natl Acad Sci USA 1999; 96:11752–11757.
Andersen JS, Lam YW, Leung AK, et al. Nucleolar proteome dynamics. Nature 2005; 433:77–83.
Das S, Chattopadhyay R, Bhakat KK, et al. Stimulation of NEIL2-mediated oxidized base excision repair via YB-1 interaction during oxidative stress. J Biol Chem 2007; 282:28474–28484.
Marsin S, Vidal AE, Sossou M, et al. Role of XRCC1 in the coordination and stimulation of oxidative DNA damage repair initiated by the DNA glycosylase hOGG1. J Biol Chem 2003; 278:44068–44074.
Parlanti E, Locatelli G, Maga G, Dogliotti E . Human base excision repair complex is physically associated to DNA replication and cell cycle regulatory proteins. Nucleic Acids Res 2007; 35:1569–1577.
Nagelhus TA, Haug T, Singh KK, et al. A sequence in the N-terminal region of human uracil-DNA glycosylase with homology to XPA interacts with the C-terminal part of the 34-kDa subunit of replication protein A. J Biol Chem 1997; 272:6561–6566.
Otterlei M, Warbrick E, Nagelhus TA, et al. Post-replicative base excision repair in replication foci. EMBO J 1999; 18:3834–3844.
Parker A, Gu Y, Mahoney W, et al. Human homolog of the MutY repair protein (hMYH) physically interacts with proteins involved in long patch DNA base excision repair. J Biol Chem 2001; 276:5547–5555.
Das A, Boldogh I, Lee JW, et al. The human Werner syndrome protein stimulates repair of oxidative DNA base damage by the DNA glycosylase NEIL1. J Biol Chem 2007; 282:26591–26602.
Boldogh I, Milligan D, Lee MS, et al. hMYH cell cycle-dependent expression, subcellular localization and association with replication foci: evidence suggesting replication-coupled repair of adenine:8-oxoguanine mispairs. Nucleic Acids Res 2001; 29:2802–2809.
Wilson SH, Kunkel TA . Passing the baton in base excision repair. Nat Struct Biol 2000; 7:176–178.
Sarker AH, Ikeda S, Nakano H, et al. Cloning and characterization of a mouse homologue (mNthl1) of Escherichia coli endonuclease III. J Mol Biol 1998; 282:761–774.
Thayer MM, Ahern H, Xing D, Cunningham RP, Tainer JA . Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J 1995; 14:4108–4120.
Liu X, Roy R . Truncation of amino-terminal tail stimulates activity of human endonuclease III (hNTH1). J Mol Biol 2002; 321:265–276.
Doublie S, Bandaru V, Bond JP, Wallace SS . The crystal structure of human endonuclease VIII-like 1 (NEIL1) reveals a zincless finger motif required for glycosylase activity. Proc Natl Acad Sci USA 2004; 101:10284–10289.
Li X, Romero P, Rani M, Dunker AK, Obradovic Z . Predicting Protein Disorder for N-, C-, and Internal Regions. Genome Inform Ser Workshop Genome Inform 1999; 10:30–40.
Marenstein DR, Ocampo MT, Chan MK, et al. Stimulation of human endonuclease III by Y box-binding protein 1 (DNA-binding protein B). Interaction between a base excision repair enzyme and a transcription factor. J Biol Chem 2001; 276:21242–21249.
Krueger KE, Srivastava S . Posttranslational protein modifications: current implications for cancer detection, prevention, and therapeutics. Mol Cell Proteomics 2006; 5:1799–1810.
Seet BT, Dikic I, Zhou MM, Pawson T . Reading protein modifications with interaction domains. Nat Rev 2006; 7:473–483.
Muller S, Hoege C, Pyrowolakis G, Jentsch S . SUMO, ubiquitin's mysterious cousin. Nat Rev 2001; 2:202–210.
Hardeland U, Steinacher R, Jiricny J, Schar P . Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J 2002; 21:1456–1464.
Bhakat KK, Mokkapati SK, Boldogh I, Hazra TK, Mitra S . Acetylation of human 8-oxoguanine-DNA glycosylase by p300 and its role in 8-oxoguanine repair in vivo. Mol Cell Biol 2006; 26:1654–1665.
Bhakat KK, Hazra TK, Mitra S . Acetylation of the human DNA glycosylase NEIL2 and inhibition of its activity. Nucleic Acids Res 2004; 32:3033–3039.
Tini M, Benecke A, Um SJ, Torchia J, Evans RM, Chambon P . Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol Cell 2002; 9:265–277.
Ramana CV, Boldogh I, Izumi T, Mitra S . Activation of apurinic/apyrimidinic endonuclease in human cells by reactive oxygen species and its correlation with their adaptive response to genotoxicity of free radicals. Proc Natl Acad Sci USA 1998; 95:5061–5066.
Das A, Hazra TK, Boldogh I, Mitra S, Bhakat KK . Induction of the human oxidized base-specific DNA glycosylase NEIL1 by reactive oxygen species. J Biol Chem 2005; 280:35272–35280.
Acknowledgements
This article with a limited focus was not meant to provide a comprehensive review of BER. Because of this and also space limitation, many appropriate references could not be included, for which the authors apologize. The authors' research is supported by US Public Health Services Research grants R01 CA81063, R01 CA53791, (SM) R01 CA102271 (TKH), P01 CA92584 and P30 ES06676. The authors thank the Mitra Lab Members for scientific discussion and Ms Wanda Smith for expert secretarial assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hegde, M., Hazra, T. & Mitra, S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res 18, 27–47 (2008). https://doi.org/10.1038/cr.2008.8
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2008.8
Keywords
This article is cited by
-
Redox dysregulation as a driver for DNA damage and its relationship to neurodegenerative diseases
Translational Neurodegeneration (2023)
-
Regulation of base excision repair during adipogenesis and osteogenesis of bone marrow-derived mesenchymal stem cells
Scientific Reports (2023)
-
DNA strand breaks and gaps target retroviral intasome binding and integration
Nature Communications (2023)
-
Biological role of the major AP (abasic site) endonuclease of an archaeon from geothermal environments
Extremophiles (2023)
-
Of the many cellular responses activated by TP53, which ones are critical for tumour suppression?
Cell Death & Differentiation (2022)