Abstract
Amycolatopsis mediterranei is used for industry-scale production of rifamycin, which plays a vital role in antimycobacterial therapy. As the first sequenced genome of the genus Amycolatopsis, the chromosome of strain U32 comprising 10 236 715 base pairs, is one of the largest prokaryotic genomes ever sequenced so far. Unlike the linear topology found in streptomycetes, this chromosome is circular, particularly similar to that of Saccharopolyspora erythraea and Nocardia farcinica, representing their close relationship in phylogeny and taxonomy. Although the predicted 9 228 protein-coding genes in the A. mediterranei genome shared the greatest number of orthologs with those of S. erythraea, it was unexpectedly followed by Streptomyces coelicolor rather than N. farcinica, indicating the distinct metabolic characteristics evolved via adaptation to diverse ecological niches. Besides a core region analogous to that common in streptomycetes, a novel 'quasi-core' with typical core characteristics is defined within the non-core region, where 21 out of the total 26 gene clusters for secondary metabolite production are located. The rifamycin biosynthesis gene cluster located in the core encodes a cytochrome P450 enzyme essential for the conversion of rifamycin SV to B, revealed by comparing to the highly homologous cluster of the rifamycin B-producing strain S699 and further confirmed by genetic complementation. The genomic information of A. mediterranei demonstrates a metabolic network orchestrated not only for extensive utilization of various carbon sources and inorganic nitrogen compounds but also for effective funneling of metabolic intermediates into the secondary antibiotic synthesis process under the control of a seemingly complex regulatory mechanism.
Similar content being viewed by others
Introduction
Amycolatopsis mediterranei is a Gram-positive filamentous actinomycete, belonging to the order of Actinomycetales and the Pseudonocardiaceae family. It produces an important antibiotic, rifamycin, whose derivatives are particularly effective against pathogenic mycobacteria, that is, Mycobacterium tuberculosis and Mycobacterium leprae 1. The medical importance of rifamycin has fostered intensive research into its biogenesis 2, 3, physiology 4, 5, and genetic manipulation 6, 7, 8, as well as characterization of the rifamycin biosynthetic gene cluster (rif) from a rifamycin B-producing strain S699 9. Although these efforts have significantly increased the strain productivity, the global reemergence of tuberculosis and the increasing emergence of rifamycin-resistant M. tuberculosis clinical strains have challenged research toward the fundamental improvement of this important ansamycin antibiotic—from its productivity to novel derivative discovery and/or design. Genome-scale information becomes critical in pursuing such research directions.
Here, we present the complete genome sequence of A. mediterranei U32, which produces rifamycin SV, biologically a much more active antibiotic than rifamycin B, and compare this sequence with those of closely related actinomycetes. By intergrating information from physiology, biochemistry and molecular biology, knowledge about the structure and function of the U32 genome has revealed the genetic basis for the phylogeny of Amycolatopsis within the Actinomycetales order, as well as the biogenesis of antibiotics, the networking of primary and secondary metabolisms, and the related probable mechanisms of regulation.
Results
General features of A. mediterranei genome
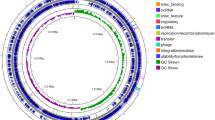
Unlike the linear topology of the chromosomes of Streptomyces coelicolor 10 and Streptomyces avermitilis 11, but resembling that of Nocardia farcinica 12 and Saccharopolyspora erythraea 13, the toplogy of the A. mediterranei chromosome is circular (Figure 1). This appears to be a common feature for the genus Amycolatopsis, because this circular topology was also suggested for the Amycolatopsis orientalis chromosome 14. With a total length of 10 236 715 base pairs (bps) (Table 1, GenBank accession number CP002000), the A. mediterranei genome is larger than that of S. coelicolor and S. erythraea, but smaller than that of the myxobacterium Sorangium cellulosum 15, and is apparently one of the largest prokaryotic genomes sequenced so far. Without any dissociated plasmids, two putative integrated plasmids with their genomic coordinates of 367 542–390 874 bp and 6 808 937–6 829 319 bp were recognized in the chromosome. Both of them were highly similar to pMEA100, which has been characterized in several species of Amycolatopsis 16. The replication origin (oriC) of the chromosome is the same as that previously characterized 7, and in this study the adjacent dnaA gene was chosen to be the starting point for numbering the 9 228 predicted protein-coding sequences (CDSs). We annotated 52 tRNA-encoding genes, with one selenocysteine tRNA (tRNASec) located at the immediate downstream of selBA, transcribed in the same direction; but upstream of selD, transcribed in the opposite direction. In addition, we also found two genes (Supplementary information, Table S1) both coding for the formate dehydrogenase α subunit, equipped with a selenocysteine (Sec)-encoding UGA codon, a Sec insertion sequence (SECIS) element, and a stem-loop structure, required for the incorporation of Sec into proteins 17. On the basis of on BLASTCLUST analysis, nearly half of the predicted CDSs (4 607, 49.9%) are clustered into 1 004 families, with membership ranging from 2 to 128 per family (Supplementary information, Table S2).
Schematic representation of the A. mediterranei chromosome and gene clusters for secondary metabolism. The outer scale is numbered in megabases and indicates the core (red), quasi-core (orange), and non-core (sky blue). Circles 1 and 2: (from the outside in), genes (forward and reverse strand, respectively) color-coded according to COG function categories. Circle 3: selected essential genes (cell division, replication, transcription, translation, and amino acid metabolism. The paralogs of essential genes in non-core are not included). Circle 4: secondary metabolic clusters—further enlarged outside the circle for detailed illustration. The colors of the key enzymes are the same as in circle 4, that is, light coral, nrps; lawn green, pks; purple, hybrid nrps and pks (n_p); plum, terpene (tps); tomato, lycopene (lyc); orange, β-carotene (car); except for the NRPS/PKS hybrid which is represented by the hybrid of light coral and lawn green (n_p); while light blue is for other function-related genes such as acp, clf, cyc, kr, and ks, that is, genes encoding acyl carrier protein, chain length factor, cyclase, β-ketoacyl reductase, and β-ketoacyl synthase, respectively. The rif cluster is further illustrated in Figure 4. Circle 5: RNAs (blue, tRNA; red, rRNA). Circle 6, mobile genetic elements (brow, transposase; light green, phage; cyan, integrated plasmids; circle 7, GC content; circle 8, GC bias (red, values > 0; blue, values < 0).
Similar to the cases of S. erythraea and S. coelicolor, an ancestral core 10, 13 containing a majority of the essential genes (Figure 1) is found to extend unequally from either side of the oriC (1.7 Mb on the left compared to 3 Mb on the right). Interestingly, within the non-core region, from the genomic coordinates of 6.6–7.5 Mb (AMED_5997 to AMED_6830), we recognized a genomic region that apparently contains more essential genes than the adjacent non-core regions (Figures 1 and 2A), including an arginine biosynthetic cluster and unique genes encoding a DNA primase, a DNA polymerase III δ subunit, translation initiation factors IF-2 and IF-3, etc. (Supplementary information, Table S1). In addition, the coding density of ortholog genes in this region is similar to the core, but distinct from that of the non-core (Figure 2A). Furthermore, the ortholog gene order of A. mediterranei genome analyzed against that of S. erythraea and N. farcinica by broken X graphics (Figure 2B and 2C), as well as against that of other actinomycetes (Supplementary information, Figure S1), showed that this region is endowed with a good consensus similar to the core. Thus, a 'quasi-core' is designated for this special region.
Genome-wise coding property of (A) A. mediterranei chromosome and the comparative broken X plot against (B) S. erythraea and (C) N. farcinica. For all the panels, x axes represent the nucleotide scale of A. mediterranei chromosome. In (A), all the dots are calculated in a 100-kb sliding window. The pink triangles represent the coding density (left, y axis), while the yellow circles represent the absolute number of essential genes (right, y axis). The turquoise squares stand for the coding density (left, y axis) of genes having orthologs in both S. erythraea and N. farcinica. The red bars represent core region. The quasi-core is highlighted in light green, and the lower coding regions in purple. In (B and C), dots represent a reciprocal best match by BLASTP comparison. Pairs of orthologs on the same strand are designated as red dots, whereas those on the opposite strand are blue.
The coding density of all CDSs is almost uniform across the chromosome except at the upstream (6.2–6.6 Mb) of the quasi-core, where it is significantly lower (77.9%) than the average (89.3%) and reaches its lowest level (66.7%) around 6.5 Mb (Figure 2A). Several transcriptional regulators, a phage-related integrase, and many hypothetical but few functional genes are found in this region, indicating probable integration of a 100-kb exogenous phage nucleotide. Furthermore, genes encoding six transposase remnants and three integrases/recombinases are unusually aggregated at the upstream of this phage-like region. This systematic variation in coding densities may imply a speed discrepancy in recombination 10 and thus we assume that the quasi-core was transferred from the core into an integration hotspot in the non-core through a transposable element-induced genomic rearrangement.
In contrast to most essential genes, genes of certain clusters of orthologous group (COG) are more focused in the non-core than in the core/quasi-core (Supplementary information, Table S3). It is particularly interesting to note that 9.73% of the genes in the non-core are involved in transcriptional regulation, whereas only 8.56% of the genes in the core/quasi-core fall into this category. Although 63 sigma factors are located in the core/quasi-core, twofold higher than that in the non-core, the numbers for anti-sigma factor and anti-anti-sigma factors are surprisingly reversed, with only eight in the core/quasi-core but 28 in the non-core. Considering the fact that similar distribution bias is also found in categories of signal transduction, lipid, carbohydrate, and secondary metabolisms (Figure 1, Supplementary information, Table S3), we postulate that the expansion of the non-core, which likely resulted mainly via adaptation to the highly competitive and complex soil environment 10, might involve not only the genes encoding auxiliary metabolic functions but also their related regulators, as shown in this study.
The phylogenetic/taxonomic characteristics of Amycolatopsis
A. mediterranei was originally classified as Sreptomyces mediterranei in 1957 18. However, later in 1969, the suggestion was made that it should have been labeled Nocardia 19 because of its cell-wall composition of meso-diaminopimilic acid (meso-DAP) and arabinose with a lack of glycine (Supplementary information, Table S4). Correspondingly, we noted that proteins known to be responsible for recruiting five glycine residues cross-bridging the peptidoglycan lateral chains 20 have been recognized in the genome of S. coelicolor (SCO0602, SCO3593 and SCO3904), but neither in A. mediterranei nor in the closely related N. farcinica and S. erythraea. Furthermore, genes involved in incorporating arabinose into the cell wall, characterized in Mycobacterium 21, can be found in the genomes of A. mediterranei, N. farcinica, and S. erythraea, but not in most of the Streptomyces species (Supplementary information, Table S5). In addition, the MurE ligase, which ligates specific amino acids 22 to the lateral chain of peptidoglycan, was analyzed phylogenetically within actinomycetes (Supplementary information, Figure S2). The MurE of A. mediterranei, and of the closely related S. erythraea, was clustered within a big clade composed of the meso-DAP-containing actinomycetes, strongly supporting the taxonomic character of A. mediterranei, that is, that meso-DAP rather than LL-DAP is used as the substrate to synthesize its cell wall.
However, by virtue of lacking mycolic acid, the most characteristic component of the cell wall in Mycobacterium and related genera, and by the inability to be infected with Nocardia-specific phages, a new genus, Amycolatopsis, was suggested in 1986 for this group of taxonomic species, including A. mediterranei 23. The studies of mycolic acid biosynthesis in M. tuberculosis show that formation of this cell wall constituent is catalyzed by at least 28 proteins 24. Critical to this process are polyketide synthase (PKS) 13 and related proteins, that is, the acyl-AMP ligase (FadD32) and the acyl-CoA carboxylase β chain (AccD4), all of which are involved in the formation of β-keto-α-alkyl mycolic acid precursors 25, 26. By analysis of 43 mycolic acid-producing actinomycetes, we found that these three proteins were clustered in a similar gene order with the same transcription directions. In addition, all PKS13 proteins demonstrate a highly conserved domain organization, that is, acyl carrier protein (ACP)-ketosynthase (KS)-acyltransferase (AT)-ACP-thioestease (TE) (Figure 3). As expected, this highly conserved cluster is not found in the genomes of actinomycetes that do not produce mycolic acids, including A. mediterranei and the closely related S. erythraea (Figure 3). In U32, two similar clusters (pks1 and pks3) were identified. However, the PKS protein in the pks1 has two additional domains (dehydratase (DH)-ketoreductase (KR)) inserted between the AT and ACP domains. The accD4 is missing in the pks3 cluster, although the corresponding PKS protein does have the same domain structure as that of M. tuberculosis. Actually, with several putative transporters and regulators annotated in these two clusters, they are more likely to code for the biosynthesis of unknown secondary metabolites (Figure 1). In addition, a bacterial type-I fatty acid synthase gene (fas-I) was found in all the mycolic acid-producing species, but absent in A. mediterranei and many of the mycolic acid-negative actinomycetes (Figure 3).
Genetic organization of the fadD-pks-accD and fas-I gene clusters in 19 selected actinobacterial genomes. The result was obtained by BLASTP using FadD32-PKS13-AccD4 and Fas-I of M. tuberculosis H37Rv as the query sequences. A total of 11 strains above the gray bar are mycolic acid-containing bacteria that harbor both fadD-pks-accD clusters and fas-I genes. The remaining seven strains lack the mycolic acids in their cell envelope. The genomes used are as follows: Corynebacterium glutamicum ATCC 13032 (NC_003450), Corynebacterium diphtheriae NCTC 13129 (NC_002935), M. tuberculosis H37Rv (NC_000962), Mycobacterium bovis AF2122/97 (NC_002945), Mycobacterium smegmatis str. MC2 155 (NC_008596), M. leprae TN (NC_002677), Mycobacterium avium 104 (NC_008595), N. farcinica IFM 10152 (NC_006361), Rhodococcus jostii RHA1 (NC_008268), Gordonia bronchialis DSM 43247 (NC_013441), Tsukamurella paurometabola DSM 20162 (NZ_ABVA00000000), Corynebacterium kroppenstedtii DSM 44385 (NC_012704), Corynebacterium amycolatum SK46 (NZ_ABZU00000000), Bifidobacterium adolescentis ATCC 15703 (NC_008618), S. avermitilis MA-4680 (NC_003155), S. erythraea NRRL 2338 (NC_009142), Frankia alni ACN14a (NC_008278).
In conclusion, the taxonomic characteristics of Amycolatopsis that relate it to but differentiate it from Streptomyces or Nocardia are intrinsically determined by their molecular phylogeny. The 16S rRNA-based phylogeny (Supplementary information, Figure S3) indicated that, S. erythraea was the closest species to A. mediterranei, followed by N. farcinica. However, although A. mediterranei shares the highest number of orthologs with S. erythraea (3 341), as expected, it is not followed by N. farcinica (2 600) but rather by the two Streptomyces species, that is, S. coelicolor (3 084) and S. avermitilis (3 068). In addition, distribution of the clusters of COG for each of the five actinomycete species demonstrated that A. mediterranei presented some focused functional clusters, particularly in transcription, signal transduction, carbohydrate transport, and metabolism, at a similar level to those in S. coelicolor, but at a higher level than those in S. erythraea, N. farcinica, and M. tuberculosis (Supplementary information, Figure S4). These apparent discrepancies may be accounted for by the fact that A. mediterranei shares the characteristic of large genome size and similar environmental conditions with that of S. coelicolor. On the other hand, taken the quantitative colinearity of orthologs into consideration, the chromosome of A. mediterranei exhibited lower values against those of S. erythraea (0.453) and N. farcinica (0.574) than that of S. coelicolor (0.620) and S. avermitilis (0.618), consistent with the 16S rRNA phylogeny. Combining the strategies of orthologs' order and genome content similarity in this study, it seems that, in terms of analyzing the relationship among actinomycetes, structural information, either from genomics or biochemistry, may better present their phylogeny while functional information such as the COG categories, particularly those related to environmental adaptation, may better present their ecology.
Potential genes for secondary metabolism and antibiotic resistance
Like the gene cluster for erythromycin synthesis (ery) in S. erythraea 13, the rif cluster of the A. mediterranei chromosome is also localized in the core. Most of the rif genes are encoded on the leading strand of replication (Figure 1), implicating this cluster as an important component of the genome critical for host survival 27.
The complete genome sequence of A. mediterranei U32 reveals at least 25 other gene clusters for biosynthesis of as-yet-uncharacterized polyketides, nonribosomal peptides, hybrid nonribosomal peptide-polyketides, and terpenoids. Only four of these clusters (rif, tps1, lyc, and nrps11) are located in the core, with one (nrps10) in the quasi-core and the other 21 scattered in the non-core (Figure 1). Besides the rif cluster, there are other four type-I and two type-II PKS clusters (Supplementary information, Table S6). In pks1 and pks3, the encoded acyl-CoA synthetases (AMED_3367 and AMED_4483) are expected to transfer an acyl starter unit to the first ACP domain of PKS1-1 and PKS3-1 proteins to synthesize their respective diketide products 28. The pks4 cluster seems to encode the synthesis of an unknown polyunsaturated fatty acid 29. PKS6-1 is highly homologous with the PKS proteins of Streptomyces globisporus 30 (52% identities) and A. orientalis 31 (79% identities), indicating that the pks6 cluster might be involved in the synthesis of an enediyne antitumor agent. The type-II pks5 cluster could be involved in the synthesis of a cyclic aromatic polyketide, because it contains both a minimal PKS unit (KS, CLF, and ACP) and two cyclases 32. There is no type-III PKS found in the genome.
Of the 11 nonribosomal peptide synthetases (NRPSs) and 4 hybrid NRPS-PKS clusters annotated in the A. mediterranei U32 genome (Supplementary information, Table S7), nrps6 appears to be involved in siderophore production, as it contains genes encoding iron-siderophore recognition- and transport-related proteins. Other clusters seem to produce completely novel secondary metabolites.
The A. mediterranei U32 genome presents a nonmevalonate pathway for generating the key C-5 precursors in terpenoid biosynthesis. Of the related four gene clusters, tps1-encoded AMED_1325 shows high end-to-end similarity to the S. coelicolor A3(2) SCO6073 (58% identities), essential for the synthesis of geosmin 33. Therefore, it is probably responsible for the synthesis of this sesquiterpene soil odor. The lyc and car seem to govern the synthesis of the antioxidant pigments lycopene and β-carotene, respectively. Scattered elsewhere in the genome, there are 55 putative cytochrome P450 enzymes (Supplementary information, Table S1), which often modify special functional groups of secondary metabolites or detoxify xenobiotics.
The A. mediterranei U32 genome contains at least 86 antibiotic-resistant genes (Supplementary information, Table S1), of which resistance to 22 antibiotics of 6 categories was experimentally verified (Supplementary information, Table S8). Unlike the non-core-focused allocation of the secondary metabolism-related gene clusters, these antibiotic-resistant genes are evenly distributed along the chromosome, indicating their essential function in conferring the ability to adapt to different hostile environments 34. As characterized in A. mediterranei S699, U32 probably employs the same two alternatives to cope with rifamycin cytotoxicity, that is, several mutations in RNA polymerase β-subunit (AMED_0656) to lower its affinity for rifamycins and a rifamycin exporter RifP (AMED_0633) to prevent the intracellular accumulation of rifamycins 1, 35.
A putative P450 enzyme (AMED_0653) is essential for the conversion of rifamycin SV to B
The 3-amino-5-hydroxybenzoic acid (AHBA) is known to be the starter unit, and two molecules of malonyl-CoA and eight molecules of (S)-methylmalonyl-CoA are extender units to form the initial macrocyclic intermediate proansamycin X 1. Several tailoring reactions, such as hydroxylation, acetylation, and methylation, are required to form the highly potent rifamycin SV 1. However, the last step, which converts rifamycin SV to the modestly active rifamycin B, is yet to be determined 36. We identified 41 single-nucleotide variations (SNVs) and 8 insertion/deletions that might affect 13 CDSs and 2 intergenic regions within the rif cluster by comparing the sequences of U32 vs. S699 (Supplementary information, Table S9). These variations were further compared to the corresponding loci of the other two rifamycin B-producing strains, A. mediterranei ATCC21789 and ATCC13685. Integration of the information of sequence comparative analysis and gene function studies 1, 36 revealed only one candidate: an asynonymous SNV leading to a missense mutation in the corresponding residue 84 of the AMED_0653 (W84) encoding a putative cytochrome P450 protein. Transforming U32 with a cloned ATCC21789 rif16 (R84) gene (pDXM4-P450), indeed led U32 to produce a high ratio of rifamycin B, with little rifamycin SV detected (Figure 4). However, without the selection pressure of apramycin in the medium, the pDXM4-P450 transformed U32 not only produced large amounts of rifamycin B but also significant amounts of rifamycin SV, likely because of the instability of the plasmid vector (Supplementary information, Figure S5). Although this function of rif16 in S699 was previously noticed via gene disruption, the mutant presented a mixture of rifamycin SV and B 36, and no complementation tests were reported. In this study, the missense mutant cytochrome P450 (W84) encoded by AMED_0653 can clearly be complemented by the prototype rif16 (R84) for conversion of rifamycin SV to B. Thus, we have provided a piece of unequivocal evidence to support the essential function of this P450 (R84). However, whether the conversion is catalyzed by this putative P450 enzyme alone, or together with the transketolases AMED_0651 and AMED_0652 as previously proposed 37, remains an open question for future analysis.
Rifamycin gene cluster (A) and sequence variation of the rif16 (AMED_0653) encoded P450 in four A. mediterranei strains and its influence on rifamycin production (B). The upper right of panel B: alignment of the Rif16 sequences from ATCC13685, ATCC21789, S699, and U32. Predicted secondary structures are shown above. Red arrow indicates the mutated residues W84 in AMED_0653, while the black arrow indicates the S699-specific amino acid variation unrelated to the conversion of rifamycin SV to B, the amino acid residue coordinates marked underneath the alignment present the positions for AMED_0653 rather than Rif16 (Orf16 for S699, refer to Supplementary information, Table S10). The center of panel B: base peak chromatograms (BPC) of rifamycin production profiles of the rifamycin B-positive controls: ATCC13685 (green), ATCC21789 (skyblue); the rifamycin SV-positive control: U32 (orange); the transformants: U32 (pDXM4-P450) (red) and the experiment negative control: U32 (pDXM4) (blue). The left of panel B: MS spectra of the rifamycin B (from ATCC21789), SV, and S (from U32). The spectrum analysis experiment was repeated for five independent U32 (pDXM4-P450) transformants.
Primary metabolism and precursors for secondary metabolite biosynthesis
As a soil inhabitant, the nutritional environment of A. mediterranei is similar to most streptomycetes, that is, rich in carbon sources but poor in nitrogen supply 38. The genomic information revealed that it could use a wide range of carbohydrates, including chitin, cellulose, xylan, and diverse oligo-/mono-saccharides (Figure 5). These carbon sources or their hydrolysates could be transported into the cell via phosphotransferase systems, ATP-binding cassette transport systems, and major facilitator superfamily transporters (Supplementary information, Table S1). Distinct from S. erythraea, N. farcinica, and S. coelicolor, U32 has a gene cluster encoding the L-arabinose isomerase, L-ribulose-5-phosphate 4-epimerase, and L-ribulokinase (AMED_4402-AMED_4404) and thus, arabinose could be converted to xylulose-5P and then enter the pentose phosphate shunt. In addition to sugars, short chain fatty acids, such as acetate and propionate, can also be consumed through the catalysis of at least four acetyl-CoA synthetases and one propionyl-CoA synthetase (Supplementary information, Table S1) to form acetyl-CoA and propionyl-CoA, respectively. Possibly, there exists an alternative pathway in U32 for the conversion of acetate to acetyl-CoA through acetaldehyde catalyzed by aldehyde dehydrogenase and acetaldehyde dehydrogenase (Supplementary information, Table S1). However, different from S. coelicolor and M. tuberculosis, if the concentration of acetate is high, A. mediterranei U32 may not be able to activate acetate through acetyl phosphate because of the lack of phosphate acetyltransferase (Pta).
Overview of carbon and nitrogen metabolic pathways and their relationships with the biosynthesis of secondary metabolites. Solid arrow, one-step reaction; two overlapped arrows, more than one-step reaction; open arrow, transmembrane transportation; dashed arrow, steps need to be further confirmed; red arrow, pathway of precursors entered into rifamycin biosynthesis; purple line, GlnR positively (+) or negatively (−) regulates the genes related to nitrogen assimilation. The color ring located at the lower left corner represents an NRPS containing the condensation (C), adenylation (A), thiolation (T), epimerization (E), and thioesterase (TE) domain. Abbreviations: Glc, glucose; Mal, maltose; GlcNAc, N-acetylglucosamine; ChB, chitobiose; Man, mannose; Fru, fructose; Rha, rhamnose; G3P, glyceraldehyde 3-phosphate; PEP, phosphoenolpyruvic acid; aminoDAHP, 3,4-dideoxy-4-amino-D-arabino-heptulosonic acid 7-phosphate; AHBA, 3-amino-5-hydroxybenzoic acid; Ru-5P, ribulose 5-phosphate; Xu-5P, xylulose 5-phosphate; Ara, arabinose; Xyl, xylose; GS, glutamine synthetase; GOGAT, glutamate synthase (NADPH); AAC, aminoglycoside N-acetyltransferase; APH, aminoglycoside O-phosphotransferase.
In addition to producing energy and reducing force, primary metabolism provides not only intermediates essential for the synthesis of cell constituents but also precursors widely used in secondary metabolism. There are at least four sets of genes encoding the acetyl-CoA and propionyl-CoA carboxylase complexes (Supplementary information, Table S1), which are used to provide malonyl-CoA and (S)-methylmalonyl-CoA for the synthesis of fatty acids and polyketides, particularly rifamycin 1, 39. An alternative pathway to generate (S)-methylmalonyl-CoA is catalyzed by methylmalonyl-CoA mutases (Supplementary information, Table S1), converting succinyl-CoA to (R)-methylmalonyl-CoA and then to (S)-isomers by methylmalonyl-CoA epimerase 40. In addition, a predicted phosphoglucomutase (AMED_0906) might be involved in the conversion of glucose-6-phospate to glucose-1-phospate, followed by the subsequent synthesis of UDP-glucose, an important precursor of AHBA 41.
The only nitrogen atom in the AHBA moiety of rifamycin was acquired from glutamine 41, and the yield of rifamycin SV was remarkably increased by as much as 171% after the addition of nitrate into the fermentation medium 4, known as the 'nitrate stimulating effect'. In U32, nitrate is firstly reduced to nitrite and then to ammonium catalyzed by the enzymes encoded by the recently characterized nasACKBDEF operon (AMED_1121-AMED_1127; Shao Z, submitted manuscript under revision). Following the sequential reactions catalyzed by glutamine synthetase (GS) and glutamate synthase, ammonium is eventually incorporated into amino acids 4 (Supplementary information, Figure S5). There are a total of six genes encoding putative type-I GSs, including the characterized glnA (AMED_1229) 42, 43 and five glnA-like genes homologous to the functionally disproved three glnA-like genes in S. coelicolor 44, but no putative type-II GS-encoding genes were found (Supplementary information, Table S10). Although the gene encoding GS adenylyltransferase (glnE, AMED_1227) was identified in the chromosome of U32, as is commonly found in streptomycetes 45 and other actinomycetes 46, the mechanism of lacking reversible adenylylation-mediated posttranslational regulation of the GS activity in strain U32 47 is yet to be revealed.
The glutamate dehydrogenase (GDH) activities were less than 1% of that of alanine dehydrogenase (AlaDH) in U32 cells grown under high concentrations of ammonia 4. Therefore, AlaDH, encoded mainly by ald (AMED_7939), is responsible for catalyzing the amination of pyruvate to yield L-alanine. Given the absence of putative genes encoding L-alanine transaminase in U32, an alternative pathway is suggested, that is, the α-amino group of L-alanine may be first transferred to 2-oxoisovalerate to form L-valine catalyzed by the valine-pyruvate transaminase (AvtA, AMED_9357) and then to form L-glutamate catalyzed by the branch-chain amino acid aminotransferases (IlvE, AMED_2179 and AMED_4755) (Figure 5).
Considering the highly biased distribution of COG genes related to transcription regulation between non-core and core/quasi-core, A. mediterranei has likely developed a complex regulatory network to coordinate the expression of genes involved in primary and secondary metabolisms. In total, 1 268 genes (13.7%) are predicted to have potential regulatory functions (Supplementary information, Table S1), including two-component systems (TCSs, 89 paired and 43 unpaired), transcriptional regulators (889), sigma/anti-sigma factors (80 out of 94 sigma factors are ECF type), and serine/threonine/tyrosine protein kinases (28). Both the absolute number and the percentage of TCSs and sigma factors identified in U32 are the highest among the five compared actinomycetes (Supplementary information, Table S2).
As a global regulator for inorganic nitrogen assimilation was identified in S. coelicolor 48, GlnR (AMED_9008) is proposed to coordinate the nitrogen assimilation in U32 as well 49 (Figure 5). Under nitrogen limitation conditions, GlnR represses the expression of ald in U32 (Wang J, unpublished data), in the same way that S. coelicolor suppresses the expression of gdhA 48 but activates the expression of both the nas operon (Wang Y, unpublished data) and the glnA gene 49. Further research will be focused on defining and characterizing the A. mediterranei-specific cis-element(s) responsible for GlnR-mediated regulation in expected nitrogen metabolism-related targets, and exploring the GlnR regulon via whole genome target analysis, aiming at a thorough understanding of the mechanism of the 'nitrate stimulating effect'.
Discussion
Completion of sequencing and annotation of the A. mediterranei U32 genome is the sum of several decades' endeavor in research on rifamycin production. The systematic analysis of the first genome sequence of the genus Amycolatopsis has endorsed a genetic basis for the phylogeny/taxonomy relationship between Streptomyces and other 'rare' genera among the Actinomycetales order. It also provides us with complete genetic information regarding the biosynthesis of rifamycin. Although strain U32 has been cultivated under laboratory conditions for decades and is well adapted to experimental manipulations, its slow growth and low transformation efficiency make genetic studies of this strain extremely difficult. Therefore, the progress made by this study will certainly open up a new era for research. The characteristic physiology of A. mediterranei can now be analyzed on the basis of a systematic comparison among the genomes of related organisms. At the same time, the previously proposed regulation models can be vigorously tested within the scope of functional genomics. These efforts will definitely facilitate the improvement of rifamycin production, as well as the functional mining of other secondary metabolites for their potential applications.
Materials and Methods
Genome sequencing and assembly
A. mediterranei strain U32 was deposited in the Institute of Microbiology, Chinese Academy of Sciences designated as CGMCC 4.5720. The bacteria used for genome sequencing were isolated from a colony-purified stock of CGMCC 4.5720, and the genomic DNA was extracted directly from the expanded culture. The nucleotide sequence was determined by 454 GS FLX sequencer 50, which resulted in 801 151 reads and provided 17.2-fold coverage. Plasmid library of 6-8 kb (pSmart), fosmid library of 35-45 kb (pCC2FOS), and 110-fold coverage solid pair-end sequencing (2 × 25 bp, Applied Biosystems) were prepared to provide contig relationship. Gaps were closed by PCR products using specially designed PCR primers. Sequence assembly was performed using phred/phrap/consed package 51, 52. The final assembly contained 808 108 sequence reads, including 801 151 reads from 454 GS FLX, 2 532 from 6-8 kb insert clone ends, 2 698 from fosmid ends, 765 from the fosmid clone shotgun, and 962 from specific PCR products and primer walking. Solid reads were also used to revise the homopolymer error in 454 raw data and the low-quality (phrap score <40) bases in assembled sequence. Totally, 105 indels and 65 SNVs that formerly existed in 454 FLX results were curated by Solid data. Together with these, an estimated error rate of < 0.5 per 100 000 bases was endued to the consensus sequence. The final assembly was confirmed in terms of restriction fragment patterns from pulse-field gel electrophoresis.
Genome annotation and analysis
Putative protein-coding sequences were predicted by glimmer 3.02 53, Genemark 54, and Z-Curve 55 softwares. CDS annotation was based on the BLASTP with KEGG, NR, and CDD databases. tRNA genes were directly predicted with tRNAscan-SE v1.23 56. Orthologous proteins between U32 and other related species were defined by reciprocal BLASTP under the condition of a minimum of 30% identity and 20% length diversity. Clustering of protein families was done by BLASTCLUST under the conditions of a minimum of 30% identity and 70% length coverage. Phylogenic trees based on 16S rRNA and MurE sequences were constructed using NJ method of the MEGA package 57, and the reliability of each branch was tested by 1 000 bootstrap replications. Antibiotic resistance genes were search against ARDB database 58 using the default parameters. Genome-wide colinearity analysis between A. mediterranei and other actinomycetes was performed similar to that used in Schneiker et al. 15.
Construction of U32 (pDXM4-P450)
Using the designed primers of P450-F, 5′-CGG ATA TCG TGT CGG TGC CGT AGA T-3′ and P450-R, 5′-CGG ATA TCA CAC GTG ATG CCT CTC TGA T-3′, the AMED_0653 gene (or orf16 of the rif cluster designated in S699 and rif16 designated in the text) encoding a prototype cytochrome P450 (R84) with its own promoter region from A. mediterranei ATCC21789 was PCR amplified and cloned into the multicopy plasmid pULVK2A derivative pDXM4 8. After the cloned DNA was verified by sequencing, the recombinant plasmid pDXM4-P450 and the vector plasmid pDXM4 as a control were introduced into U32 by electroporation, as described 8.
Rifamycin production analysis
Cultures of the wild-type and its transformant strains were grown for 5 days in Benett's liquid medium (glucose 1%, tryptone 0.2%, yeast extract 0.1%, beef extract 0.1%, glycerol 1% (w/v, pH 7.0)) at 30 °C. The culture broths were adjusted to pH 2-3 by 1 M HCl and extracted once with equal volumes of ethyl acetate 36. The ethyl acetate solutions were filtered (0.22 μm) and then directly analyzed by HPLC-MS (Agilent HPLC 1200 MS Q-TOF 6520 System). HPLC was performed on a Zorbax Eclipse XDB-C18 column (50 × 4.6mm, 1.8 μm; gradient methanol: 0.5% formic acid in water at t0 = 70:30, at t15min = 90:10, at t18min = 70:30, and stop at t23min; 0.2 ml/min flow rate) with detection wavelength at 256 and 425 nm. To obtain the electrospray mass spectra of all peaks, the TIC-positive mode was employed and the mass spectrometric parameters are as follows: mass range 550-1 100 m/z (MS scan rate 1.03 and resolution ± 0.5 amu), nebulizer 40 psi, gas (N2) temperature 350 °C, gas flow 9 L/min, VCap 3500 V, Fragmentor 160 V, Skimmer 65 V, Octopole RF 750 V, and Ext Dyn standard 2 GHz (3200). The temperature of the ion spray was maintained at 21 ± 1 °C. To validate the rifamycin peaks, the MS2-positive ion mode mass spectrometry was used. All the MS2 parameters are the same as those in TIC above, except MS2 range 100-1 000 and collision energy 35 V. The fragmentation of 756 and 778 m/z ion (Rifamycin B) was monitored for the first chromatographic run at 8.411 min, that of 720 m/z ion (Rifamycin SV) was monitored for the scecod at 9.667 min, and that of 696 and 718 m/z ion (Rifamycin S, oxidative form of rifamycin SV in the air) was monitored for the third at 15.132 min.
Accession codes
References
Floss HG, Yu TW . Rifamycin-mode of action, resistance, and biosynthesis. Chem Rev 2005; 105:621–632.
White RJ, Martinelli E, Gallo GG, Lancini G, Beynon P . Rifamycin biosynthesis studied with 13C enriched precursors and carbon magnetic resonance. Nature 1973; 243:273–277.
Lal R, Khanna M, Kaur H, et al. Rifamycins: strain improvement program. Crit Rev Microbiol 1995; 21:19–30.
Chiao JS, Xia T, Mei BG, Jin ZK, Gu WL . Rifamycin SV and related ansamycins, regulation of biosynthesis. In: Vining LC, Stuttard C. eds. Genetics and Biochemistry of Antibiotic Production. Boston: Butterworth-Heinemann, 1996: 477–498.
Yang YL, Jiang WH, Chiao JS, Zhao GP . Regulation of rifamycin SV production and glutamine synthetase expression in Amycolatopsis mediterranei U-32. Actinomycetol 1998; 12:141–147.
Lal R, Lal S, Grund E, Eichenlaub R . Construction of a hybrid plasmid capable of replication in Amycolatopsis mediterranei. Appl Environ Microbiol 1991; 57:665–671.
Tian Y, Hao P, Zhao G, Qin Z . Cloning and characterization of the chromosomal replication origin region of Amycolatopsis mediterranei U32. Biochem Biophys Res Commun 2005; 333:14–20.
Ding X, Tian Y, Chiao J, Zhao G, Jiang W . Stability of plasmid pA387 derivatives in Amycolatopsis mediterranei producing rifamycin. Biotechnol Lett 2003; 25:1647–1652.
August PR, Tang L, Yoon YJ, et al. Biosynthesis of the ansamycin antibiotic rifamycin: deductions from the molecular analysis of the rif biosynthetic gene cluster of Amycolatopsis mediterranei S699. Chem Biol 1998; 5:69–79.
Bentley SD, Chater KF, Cerdeno-Tarraga AM, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 2002; 417:141–147.
Ikeda H, Ishikawa J, Hanamoto A, et al. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat Biotechnol 2003; 21:526–531.
Ishikawa J, Yamashita A, Mikami Y, et al. The complete genomic sequence of Nocardia farcinica IFM 10152. Proc Natl Acad Sci USA 2004; 101:14925–14930.
Oliynyk M, Samborskyy M, Lester JB, et al. Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nat Biotechnol 2007; 25:447–453.
Redenbach M, Scheel J, Schmidt U . Chromosome topology and genome size of selected actinomycetes species. Antonie Van Leeuwenhoek 2000; 78:227–235.
Schneiker S, Perlova O, Kaiser O, et al. Complete genome sequence of the myxobacterium Sorangium cellulosum. Nat Biotechnol 2007; 25:1281–1289.
te Poele EM, Habets MN, Tan GY, et al. Prevalence and distribution of nucleotide sequences typical for pMEA-like accessory genetic elements in the genus Amycolatopsis. FEMS Microbiol Ecol 2007; 61:285–294.
Zhang Y, Gladyshev VN . An algorithm for identification of bacterial selenocysteine insertion sequence elements and selenoprotein genes. Bioinformatics 2005; 21:2580–2589.
Margalith P, Beretta G . Rifomycin.XI. taxonomic study on Streptomyces mediterranei nov. sp. Mycopathol Mycol Appl 1960; 13:321–330.
Thiemann JE, Zucco G, Pelizza G . A proposal for the transfer of Streptomyces mediterranei Margalith and Beretta 1960 to the genus Nocardia as Nocardia mediterranea (Margalith and Beretta) comb. nov. Arch Mikrobiol 1969; 67:147–155.
Ton-That H, Labischinski H, Berger-Bachi B, Schneewind O . Anchor structure of staphylococcal surface proteins. III. Role of the FemA, FemB, and FemX factors in anchoring surface proteins to the bacterial cell wall. J Biol Chem 1998; 273:29143–29149.
Wolucka BA . Biosynthesis of D-arabinose in mycobacteria - a novel bacterial pathway with implications for antimycobacterial therapy. Febs J 2008; 275:2691–2711.
Gordon E, Flouret B, Chantalat L, et al. Crystal structure of UDP-N-acetylmuramoyl-L-alanyl-D-glutamate: meso-diaminopimelate ligase from Escherichia coli. J Biol Chem 2001; 276:10999–11006.
Lechevalier MP, Prauser D, Labeda P, Ruan J-S . Two new genera of nocardioform actinomycetes: Amycolata gen.nov. and Amycolatopsis. gen. nov. Int J Syst Bacteriol 1986; 36:29–37.
Raman K, Rajagopalan P, Chandra N . Hallmarks of mycolic acid biosynthesis: a comparative genomics study. Proteins 2007; 69:358–368.
Portevin D, De Sousa-D'Auria C, Houssin C, et al. A polyketide synthase catalyzes the last condensation step of mycolic acid biosynthesis in mycobacteria and related organisms. Proc Natl Acad Sci USA 2004; 101:314–319.
Portevin D, de Sousa-D'Auria C, Montrozier H, et al. The acyl-AMP ligase FadD32 and AccD4-containing acyl-CoA carboxylase are required for the synthesis of mycolic acids and essential for mycobacterial growth: identification of the carboxylation product and determination of the acyl-CoA carboxylase components. J Biol Chem 2005; 280:8862–8874.
Price MN, Alm EJ, Arkin AP . Interruptions in gene expression drive highly expressed operons to the leading strand of DNA replication. Nucleic Acids Res 2005; 33:3224–3234.
Trivedi OA, Arora P, Sridharan V, et al. Enzymic activation and transfer of fatty acids as acyl-adenylates in mycobacteria. Nature 2004; 428:441–445.
Metz JG, Roessler P, Facciotti D, et al. Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes. Science 2001; 293:290–293.
Liu W, Christenson SD, Standage S, Shen B . Biosynthesis of the enediyne antitumor antibiotic C-1027. Science 2002; 297:1170–1173.
Zazopoulos E, Huang K, Staffa A, et al. A genomics-guided approach for discovering and expressing cryptic metabolic pathways. Nat Biotechnol 2003; 21:187–190.
Shen Y, Yoon P, Yu TW, et al. Ectopic expression of the minimal whiE polyketide synthase generates a library of aromatic polyketides of diverse sizes and shapes. Proc Natl Acad Sci USA 1999; 96:3622–3627.
Jiang J, He X, Cane DE . Geosmin biosynthesis. Streptomyces coelicolor germacradienol/germacrene D synthase converts farnesyl diphosphate to geosmin. J Am Chem Soc 2006; 128:8128–8129.
Hopwood DA . How do antibiotic-producing bacteria ensure their self-resistance before antibiotic biosynthesis incapacitates them? Mol Microbiol 2007; 63:937–940.
Absalon AE, Fernandez FJ, Olivares PX, et al. RifP; a membrane protein involved in rifamycin export in Amycolatopsis mediterranei. Biotechnol Lett 2007; 29:951–958.
Xu J, Wan E, Kim CJ, Floss HG, Mahmud T . Identification of tailoring genes involved in the modification of the polyketide backbone of rifamycin B by Amycolatopsis mediterranei S699. Microbiology 2005; 151:2515–2528.
Floss HG . From ergot to ansamycins-45 years in biosynthesis. J Nat Prod 2006; 69:158–169.
Hodgson DA . Primary metabolism and its control in streptomycetes: a most unusual group of bacteria. Adv Microb Physiol 2000; 42:47–238.
Diacovich L, Peiru S, Kurth D, et al. Kinetic and structural analysis of a new group of Acyl-CoA carboxylases found in Streptomyces coelicolor A3(2). J Biol Chem 2002; 277:31228–31236.
Zhang W, Yang L, Jiang W, et al. Molecular analysis and heterologous expression of the gene encoding methylmalonyl-coenzyme A mutase from rifamycin SV-producing strain Amycolatopsis mediterranei U32. Appl Biochem Biotechnol 1999; 82:209–225.
Guo J, Frost JW . Kanosamine biosynthesis: a likely source of the aminoshikimate pathway's nitrogen atom. J Am Chem Soc 2002; 124:10642–10643.
Mei BG, Jiao RS . Purification and properties of glutamate synthase from Nocardia mediterranei. J Bacteriol 1988; 170:1940–1944.
Peng WT, Wang J, Wu T, et al. Bacterial type I glutamine synthetase of the rifamycin SV producing actinomycete, Amycolatopsis mediterranei U32, is the only enzyme responsible for glutamine synthesis under physiological conditions. Acta Biochim Biophys Sin 2006; 38:821–830.
Rexer HU, Schaberle T, Wohlleben W, Engels A . Investigation of the functional properties and regulation of three glutamine synthetase-like genes in Streptomyces coelicolor A3(2). Arch Microbiol 2006; 186:447–458.
Fisher SH, Wray LV, Jr . Regulation of glutamine synthetase in Streptomyces coelicolor. J Bacteriol 1989; 171:2378–2383.
Carroll P, Pashley CA, Parish T . Functional analysis of GlnE, an essential adenylyl transferase in Mycobacterium tuberculosis. J Bacteriol 2008; 190:4894–4902.
Mei BG, Chiao JS . Studies on glutamine synthetase from Nocardia mediterranei. II. Regulation of enzyme activity and some kinetic properties. Acta Biochim Biophys Sin 1986; 18:500–511.
Tiffert Y, Supra P, Wurm R, et al. The Streptomyces coelicolor GlnR regulon: identification of new GlnR targets and evidence for a central role of GlnR in nitrogen metabolism in actinomycetes. Mol Microbiol 2008; 67:861–880.
Yu H, Peng WT, Liu Y, et al. Identification and characterization of glnA promoter and its corresponding trans-regulatory protein GlnR in the rifamycin SV producing actinomycete, Amycolatopsis mediterranei U32. Acta Biochim Biophys Sin 2006; 38:831–843.
Margulies M, Egholm M, Altman WE, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005; 437:376–380.
Ewing B, Hillier L, Wendl MC, Green P . Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 1998; 8:175–185.
Gordon D, Abajian C, Green P . Consed: a graphical tool for sequence finishing. Genome Res 1998; 8:195–202.
Delcher AL, Harmon D, Kasif S, White O, Salzberg SL . Improved microbial gene identification with GLIMMER. Nucleic Acids Res 1999; 27:4636–4641.
Lukashin AV, Borodovsky M . GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res 1998; 26:1107–1115.
Guo FB, Ou HY, Zhang CT . ZCURVE: a new system for recognizing protein-coding genes in bacterial and archaeal genomes. Nucleic Acids Res 2003; 31:1780–1789.
Lowe TM, Eddy SR . tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 1997; 25:955–964.
Tamura K, Dudley J, Nei M, Kumar S . MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 2007; 24:1596–1599.
Liu B, Pop M . ARDB--antibiotic resistance genes database. Nucleic Acids Res 2009; 37:D443–D447.
Acknowledgements
This paper is dedicated to the late Professor JS Chiao, who initiated the research in China for rifamycin production employing A. mediterranei more than 30 years ago and who continued the endeavor to resolve the mechanism of the 'nitrate stimulating effect' up to the last breath of his life. This work was supported by the National Natural Science Foundation of China (30830002), the National High Technology Research and Development Program of China (2007AA021301, 2007AA021503), and the Research Unit Fund of Li Ka Shing Institute of Health Sciences (7103506).
Author information
Authors and Affiliations
Corresponding authors
Additional information
( Supplementary information is linked to the online version of the paper on Cell Research website.)
Supplementary information
Supplementary information, Table S1
Selected genes/proteins mentioned in the main text (PDF 185 kb)
Supplementary information, Table S2
A selection of protein families among five related actinobacteria. (PDF 49 kb)
Supplementary information, Table S3
Gene distribution of different functional categories. (PDF 31 kb)
Supplementary information, Table S4
General biochemical features of cell wall in related actinomytes' genera. (PDF 36 kb)
Supplementary information, Table S5
Genes involved in the pathway to incorporate arabinose into the cell wall characterized in different actinomycetes. (PDF 36 kb)
Supplementary information, Table S6
Deduced functions of PKS proteins in polyketide biosynthetic gene clusters. (PDF 40 kb)
Supplementary information, Table S7
Deduced functions of NRPS proteins in non-ribosomal peptide biosynthetic gene clusters. (PDF 46 kb)
Supplementary information, Table S8
Drug resistance profile of A. mediterranei. (PDF 34 kb)
Supplementary information, Table S9
Genetic variations of the rifamycin biosynthetic gene cluster detected in A. mediterranei strain S699 and U32. (PDF 52 kb)
Supplementary information, Table S10
BlastP results of glutamine synthetases from A. mediterranei U32. (PDF 41 kb)
Supplementary information, Figure S1
Broken X comparison of genome structure for A. mediterranei versus S. coelicolor, S. avermitilis, M. tuberculosis and Frankia alni. (PDF 200 kb)
Supplementary information, Figure S2
Pylogenetic analysis of MurE in actinomycetes. (PDF 187 kb)
Supplementary information, Figure S3
Phylogeny tree based on 16S rRNA among selected actinobacteria and other related species. (PDF 218 kb)
Supplementary information, Figure S4
COG category distributions of predicted genes in A. mediterranei, S. erythraea, N. farcinica, S. coelicolor and M. tuberculosis. (PDF 164 kb)
Supplementary information, Figure S5
The effects of pDXM4-P450 instability property on the production profiles of different kinds of rifamycin. (PDF 148 kb)
Rights and permissions
About this article
Cite this article
Zhao, W., Zhong, Y., Yuan, H. et al. Complete genome sequence of the rifamycin SV-producing Amycolatopsis mediterranei U32 revealed its genetic characteristics in phylogeny and metabolism. Cell Res 20, 1096–1108 (2010). https://doi.org/10.1038/cr.2010.87
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2010.87
Keywords
This article is cited by
-
Heterologous production of the D-cycloserine intermediate O-acetyl-L-serine in a human type II pulmonary cell model
Scientific Reports (2023)
-
Transposon-based identification of genes involved in the rimocidin biosynthesis in Streptomyces rimosus M527
World Journal of Microbiology and Biotechnology (2023)
-
Inter domain interactions influence the substrate affinity and hydrolysis product specificity of xylanase from Streptomyces chartreusis L1105
Annals of Microbiology (2020)
-
Natural product drug discovery in the genomic era: realities, conjectures, misconceptions, and opportunities
Journal of Industrial Microbiology and Biotechnology (2019)
-
Comparative genomics reveals phylogenetic distribution patterns of secondary metabolites in Amycolatopsis species
BMC Genomics (2018)