Abstract
Hereditary hemorrhagic telangiectasia (HHT) is an autosomal dominant disease characterized by widespread arteriovenous malformations and caused by mutations in two major genes: ENG and ACVRL1. Two decades ago, a French epidemiological study pointed out that its prevalence was higher than previously thought and that its distribution varied greatly from one area to another, one of the highest concentrations of patients being found in the Haut-Jura mountains. Although germline mutations are usually family specific, some of them have been reported in unrelated patients, especially for ACVRL1. We performed haplotype analysis of 116 French and Italian patients carrying 13 ACVRL1 different mutations. For five of these mutations, we estimated the age of the most recent common ancestors (MRCAs) using the ESTIAGE program. Most mutations were related to both recurrent mutational events and founder effects with age estimates ranging from 100 to 550 years. The c.1112dupG mutation, which is likely to be responsible for the very high concentration of HHT patients found in the former epidemiological study, probably occurred in one inhabitant of the Haut-Jura Mountains more than three centuries ago. The p.Arg374Gln mutation occurred independently in at least two distinct geographical areas, including the area with the second highest prevalence in the epidemiological study and where the MRCA is rather recent (about 100 years ago). Partially shared haplotypes between French and Italian patients were found for three mutations. This suggests a common origin and a possible diffusion of these mutations from Italy to France.
Similar content being viewed by others
Introduction
Hereditary hemorrhagic telangiectasia (HHT, Rendu-Osler-Weber disease) (OMIM#187300) is an autosomal dominant disease characterized by widespread arteriovenous malformations. Common manifestations are severe and recurrent nosebleeds and mucocutaneous telangiectases, which are clusters of abnormally dilated vessels. Visceral complications include pulmonary, hepatic and cerebral arteriovenous malformations and can be life threatening.1
Hereditary hemorrhagic telangiectasia is caused by germline mutations in two major genes: ENG (GenBank AH006911), encoding endoglin, a TGF-β type III receptor and ACVRL1 (GenBank AH005451), encoding activin receptor-like kinase type-1 (ALK-1), a putative TGF-β type I receptor.2, 3 ALK-1 induces the activation of Smad-1/Smad-5, which is considered to balance the ALK-5 pathway through Smad-2/Smad-3 in endothelial cells, whereas endoglin influences both ALK-1 and ALK-5 pathways.4, 5 Evidence for the existence of two other HHT-associated genes has recently been reported and mutations have also been identified in MADH4 in a subset of families with combined juvenile polyposis and HHT.6, 7, 8 Mutations of ACVRL1 and ENG account for nearly 90% of our HHT patients.9 Although mutations are found in most part of ENG and ACVRL1 and are usually private (ie, family specific), some of them have been reported in patients from unrelated families and in different countries, especially for ACVRL1.10
Hereditary hemorrhagic telangiectasia has long been underdiagnosed and considered as a rare disease. Twenty years ago, a large-scale French epidemiological study found an average prevalence of 1/8345, which was more than 10 times higher than expected at that time.11 This is consistent with more recent studies performed in different populations also showing that the disease was present worldwide.12, 13 The French epidemiological study also pointed out that the distribution of the disease could vary greatly from one area to another. Three French administrative areas had a far higher prevalence: Ain (1/3345), Jura (1/5062) and Deux-Sèvres (1/4287).11
Here, we report on the haplotype analysis of 13 ACVRL1 mutations, which have been found in different patients from France and Italy. For five of these mutations, we estimate the time of their introduction in the respective population. We also show that the c.1112dupG mutation is responsible for the very high frequency of the disease observed in the Ain and Jura administrative areas. This mutation is likely to have occurred in a common ancestor living in a valley of the Haut-Jura mountains more than three centuries ago and to have spread over the generations, mainly in the Rhône-Alpes region but also outside.
Materials and methods
Patients
A total of 96 probands from apparently unrelated nuclear HHT families, who had been referred for molecular diagnosis to one of the medical centers of the French-Italian HHT Network in France (n=89) or Northern Italy (n=7), were studied. Thirty-five of them carried the c.1112dupG mutation, whereas the 61 others carried one of the 12 different point mutations. When available, one affected or unaffected close relative was also studied to allow haplotype reconstruction (n=19). Informed written consent has been obtained from the patients according to the French and Italian Bioethics laws. For determination of allele frequencies at the different microsatellites, DNA from 20 French control individuals (40 chromosomes) was studied.
Mutation analysis
Germline mutation analysis of the index cases and their relatives has been performed in the three genetics laboratories from the French-Italian HHT networks in Lyon (n=91), Paris (n=14) and Pavia (n=10) as previously reported.9, 10, 14 The following mutations were included in the present study: p.Arg67Trp (c.199C>T), p.Arg144X (c.430C>T), c.1112dupG (p.Gly371fsX), p.Arg374Trp (c.1120C>T), p.Arg374Gln (c.1121G>A), p.Met376Val (c.1126A>G), p.Arg411Trp (c.1231C>T), p.Arg411Gln (c.1232G>A), p.Asp427Val (c.1280A>T), p.Arg479Gln (c.1436G>A), p.Arg479X (c.14355C>T), p.Ala482Val (c.1445C>T) and p.Arg484Trp (c.1450C>T).
Haplotype analysis
Haplotype analysis was performed in the genetics laboratory of Lyon. One intragenic and nine flanking polymorphic microsatellite markers spanning over 20 megabases were studied: D12S1653 (AFMB283XH5), D12S85 (AFM122XF6), D12S2196 (GATA137B11), D12S1677 (AFMB347VB9), D12S1712 (AFMA060YE9), D12S270 (MFD305), D12S262, D12S1724 (AFMA083YC9), D12S90 (AFM172XD8) and D12S83 (AFM112YF4). Markers were selected using the UCSC genome browser (http://genome.ucsc.edu/cgi-bin/hgGateway). Their order and their distances relative to ACVRL1 are shown in Figure 1. The genetic distance between D12S1653 and D12S83 is 18.79 cM. Primers used for PCR were those provided by the database except for D12S83 and D12S90, for which novel primers have been designed using the Primer3 free online software (http://www-genome.wi.mit.edu/genome_software/other/primer3.html): D12S83 forward, 5′-tttctctcatttcctgcactctc-3′; D12S83 reverse, 5′-tgaggaagatcaatgaaaaggtt-3′; D12S90 forward, 5′-gggaggatagaaaagcagca-3′; and D12S90 reverse, 5′-agtcaggcccacccaattta-3′. PCR was performed using fluorescently labeled F-primers according to standard methods (reaction conditions available on request). PCR products were loaded on an ABI Prism 377 automated sequencer and analyzed using the Genescan Analysis software (Applied Biosystems, Foster City, CA, USA).
Haplotype reconstruction and estimation of the age of the most recent common ancestors of ACVRL1 mutation carriers
To determine if founder effects were likely to exist for each mutation, five DNA microsatellites, closely linked to ACVRL1, were first studied. They included the intragenic microsatellite marker (D12S1677) and two flanking markers on each side of ACVRL1 (D12S85, D12S2196, D12S1712 and D12S270). If alleles at these closely linked markers were shared by all (or a subset of) patients carrying one given mutation, a founder effect for the mutation was suspected and a more extensive haplotype analysis was performed using five additional microsatellites spanning over 20 megabases around the ACVRL1 gene. These data were used to confirm the founder effects by identifying the most likely ancestral haplotypes associated with each of these mutations and to estimate the age of the different founder events using the ESTIAGE program.15 This latter program implements a likelihood-based method to estimate the age of the most recent common ancestor (MRCA) of a group of patients carrying a same mutation using multilocus marker data from these patients. It is assumed that all these patients descended from a common ancestor, who introduced the mutation ngen generation ago. An estimate of ngen is obtained from the size of the haplotype shared by the individuals on each side of the disease locus by finding the most likely positions of recombinations on the ancestral haplotype. Allele frequencies of the markers and mutation rates are taken into account to allow for the possibility either that recombination occurred in previous intervals with haplotypes sharing the same alleles as the ancestral haplotype, or to let haplotype diversity be due to mutations rather than recombinations. A mutation rate of 10–4 per individual per generation and a stepwise mutation model are assumed at the microsatellite markers. The most likely ancestral haplotype associated with each mutation is determined by considering markers on each side of the mutation, one at a time, starting from the mutation and sequentially identifying the most frequent allele in patients carrying the presumed ancestral allele at the previous marker. Patients are then assumed to carry the ancestral haplotype on each side of the mutation up to the first marker where they do not show an allele similar to the ancestral allele at this locus.
Geographic distribution of the HHT families in the Rhône-Alpes region
For the index cases living in the Rhône-Alpes region, we collected data on their birth places or, when not available, the town where they lived at the time of the study.
Results
Evidence of possible founder effects for several of the ACVRL1 point mutations
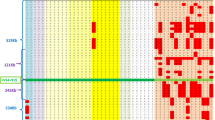
Figure 2 shows, for the 12 point mutations, the genotypes and the shared alleles of the patients at the intragenic and the four flanking markers. When DNA from close relatives was available, haplotypes were manually reconstructed. For several mutations, recurrent mutation events are suggested by the fact that no single associated haplotype are observed. However, founder effects may be associated with some of these different mutational events as allele sharing is observed in some subsets of patients. This is in particular the case for the p.Arg374Gln mutation. Three completely different haplotypes are observed in association with this mutation. For two of them, a geographical clustering of patients is observed. Indeed, haplotype 2-8-374Gln-2-4-7 is found essentially in patients living in the Deux-Sèvres area and haplotype 2-8-374Gln-6-5-? in patients from the northeast part of France. No clear regional clustering is observed for the other mutations with suspected founder effect.
Haplotypes of the patients sharing the common missense and nonsense mutations of ACVRL1. When available, the haplotype of a relative, carrying the mutation (#) or not (*), was added for phase determination. Isolate probands or families are separated by vertical lines. French patients are encoded with numerals whereas Italian patients are encoded with letters. The intragenic marker D12S1677 is underlined.
For the p.Arg67Gln and p.Arg144X mutations, the Italian patients shared a common haplotype, although they were not apparently related. For the mutation p.Arg484Trp, French and Italian patients had a completely distinct haplotype, whereas two unrelated Italian patients and several French patients shared a partial common haplotype for the p.Arg144X or the p.Arg374Trp mutations. For the p.Arg67Gln mutation, one French patient shared one allele of the intragenic D12S1677 marker with the Italian patients.
Estimation of the age of the founder effect for ACVRL1 mutations
Observed genotypes in patients carrying the c.1112dupG and the four ACVRL1 mutations exhibiting founder effects are shown in Table 1. Allele frequencies in control individuals are shown in Supplementary Table 1. For mutation p.Arg374Gln, only the first three patients from the Deux-Sèvres area are considered as different founder effects are suspected based on the analysis of the most proximal markers. The age of the MRCA of these different mutations are shown in Table 2 with their 95% confidence intervals and the presumed ancestral haplotype. The mutation p.Arg374Gln appears to be the most recently introduced one (approximately one century old, although confidence interval is large) and very similar age estimates are found for mutations c.1112dupG, p.Arg411Trp and p.Arg374Trp that are all found to be approximately 300 years old. The mutation p.Arg144X is probably the oldest one with an age estimate of about 550 years.
Geographic distribution of the HHT families in the Rhône-Alpes region
Figure 3a shows the distribution of the HHT families in the Rhône-Alpes region in 1977, many years prior to the identification of ENG or ACVRL1 genes. The highest concentration of patients was found in the Haut-Jura mountains, in a 50-km area around the Valserine Valley. Figure 3c shows, on a Rhône-Alpes-centered map, the distribution of the birth places, or when not available the present addresses, of the probands with a mutation in either ENG or ACVRL1.
(a) Living locations of the patients affected with HHT in the Rhône-Alpes region in 1977. The size of the black circles is related to the number of patients. The dotted red circle refers to the 50-km diameter area with the highest concentration of patients in that period. This area was centered on the Valserine Valley (villages of Montanges, Champfromier, Chézery-Forens and Lélex). (b) Localization of the studied area, between the cities of Lyon and Geneva (Switzerland). (c) Places of birth or present living location of the HHT probands with an ENG or ACVRL1 mutation, in an area centered on the Rhône-Alpes region. Index cases with the c.1112dupG mutation were distinguished from those with other ACVRL1 mutations and from those with ENG mutations.
Discussion
ACVRL1 mutations result from different mechanisms
The most frequent mutation found in French HHT patients is a one-basepair guanidine insertion, c.1112dupG.10 At the time of this study, it was present in 36 apparently unrelated index cases and has not been reported in other countries except for an American patient originating from the Ain administrative area.16 This mutation is certainly the consequence of a unique mutational event as shown by the fact that all the patients share a common haplotype. The age of the MRCA of these patients was estimated to be 325 years, consistent with the fact that all patients except one have not recombined at the nearest markers on both sides of the mutation and haplotype sharing extends over several megabases for several patients.
The other types of ACVRL1 mutations found in several patients consist of point mutations, which are likely to result from different independent mutational events. In all, 10 out of the 12 point mutations were the products of C>T transitions (or the reverse G>A transition) and 9 occurred on codons encoding arginine. This strongly suggests a common and recurrent mechanism by spontaneous deamination of the methyl CpG during development leading to a T and not corrected by the mismatch repair process.17 These mutations have been reported in several occurrences in different populations as recorded in the HHT mutation database (http://www.hhtmutation.org/). Codons 374 and 411 are the most frequently involved and can be considered as mutation hot spots.10 However, several apparently unrelated patients with a given mutation share the same haplotype, suggesting the occurrence of different founder effects. This is especially true for the most common mutations: p.Arg144X, p.Arg374Gln, p.Arg374Trp and p.Arg411Trp.
The remaining two missense mutations are likely to result from a less common mechanism as suggested by the fact that they have been reported less frequently: transition A>G for the p.Met376Val and transversion A>T for the p.Asp427Gln.
The classical French epidemiological study of HHT revisited
This large-scale epidemiological study was planned in 1984 after the observation of an unusually high frequency of the disease in the Haut-Jura area. It was based on a postal questionnaire sent to 23 000 physicians from 52 administrative areas (on a total of 96 for metropolitan France). An unexpectedly high frequency (1/8345) of HHT was found with large differences in prevalence between neighboring areas. At that time, HHT was considered as a very rare disease and neither its genetic heterogeneity nor the high allelic heterogeneity was predicted. In conclusion of the study, two alternative hypotheses were proposed to explain the disease diffusion: ‘were there several foci of origin, or was the pathogenic gene disseminated from a single source?’
More than 20 years later, after the identification of the two major HHT genes and the unraveling of its considerable allelic heterogeneity, it appears that data observed in the epidemiological study can be explained by the combination of both hypotheses. Our molecular study shows that many different mutations of both ENG and ACVRL1 are present in France and even in the administrative areas with a very high prevalence of the disease, like Jura and Ain.9, 10 However, a founder effect associated to the c.1112dupG mutation is responsible for the higher concentration of HHT patients in this area, compared with the rest of the country.
Genealogical studies performed on the HHT families originating from the Valserine Valley and the surrounding Haut-Jura mountains showed that these patients shared a few common ancestors.18 However, it was not possible to identify a single common ancestor that would have introduced the mutation, as the church archives used to reconstruct genealogies were not collected prior to the seventieth century (the province corresponding to most of the present Ain administrative area was transferred to the French kingdom from the Duchy of Savoy, by the treaty of Lyon, in 1601). Combining our results with data from the genealogical studies, we can hypothesize that the present distribution of the c.1112dupG mutation may result from three successive mechanisms.18 First, this unique mutation may have occurred prior to the seventieth century in one inhabitant from the Valserine Valley or from the neighboring areas. Second, its frequency would have progressively increased due to genetic drift. Although the population of this valley cannot be considered as genetically isolated, since continuous immigration and emigration flows have been documented, migrants were shown to have a relatively small contribution to the present population, which is mainly related to a small nucleus of families, stable over centuries.18 Third, the mutation began to spread in the ninetieth century to the close industrializing towns and next to major cities within and outside the Rhône-Alpes region. Since this migration process was still ongoing during the last decades, it is not unlikely that the contrasting HHT frequency in Jura and Ain and in the other administrative areas could be less pronounced, would the epidemiological study be performed today rather than one generation ago.
Other founder effects associated with given geographic areas
The haplotype 2-8-374Gln-2-4-7 of the p.Arg374Gln mutation has been found in patients living in the Deux-Sèvres administrative area, suggesting a founder effect that can also be linked to the high prevalence of the disease observed in this area in the former epidemiological study.11 Estimates of the age of the MRCA of these patients are in favor of a recent introduction of this mutation in the population (100 years), but due to the limited number of index cases available for the study, confidence interval is rather large (from 50 to 400 years). However, this is consistent with its restricted geographical distribution and with the fact that the concentration of HHT patients was lower than in the Haut-Jura area. The contrast with the lack of obvious genealogical evidence for a common ancestor may be due to the fact that patients have been recruited on the basis of a medical genetics consultation and that their extensive genealogy could not be raised up to a few generations. Consequently, a common ancestor living earlier than several generations ago, which is not unlikely due to the rather wide confidence interval, would not be easily identified. Interestingly, a distinct haplotype for the same mutation (2-8-374Gln-6-5-?) was found in three patients from the northeast part of France, suggesting a distinct founder effect.
The p.Arg411Trp and p.Arg374Trp mutations associated with the ancestral haplotypes 2-2-8-2-411Trp-4-2-6-7-8-10 and 2-2-8-2-374Trp-4-2-3-1-7-9, respectively, and both estimated to have been introduced in the population about 300 years ago, were not found to be linked to any given French geographic area. Neither was the p.Arg144X mutation, associated with the ancestral haplotype 8-8-6-5-144X-6-7-8-3-?-?, which was estimated to have been introduced earlier (about 500 years ago).
In the literature, two founder effects have been reported, both involving the ENG gene. The p.Tyr120X (c.360C>A) mutation was found in seven apparently unrelated families living in the Funen county of Denmark. Haplotype analysis was in favor of a founder effect, and the introduction of the mutation in the population was estimated to have occurred about 350 years ago.19 The c.67+1G>A mutation has been found in seven families from the Leeward Islands of the Dutch West Indies that have a very high prevalence of HHT (1/1639). This mutation has also been found in a patient living in the Netherlands and haplotype analysis suggested its probable introduction in these islands, which were mainly populated by descendants of African slaves, by a Dutch settler.20
Recurrent ACVRL1 mutations and migrations from Italy to France
France has been a country of immigration from different European countries for centuries. Since a continuous emigration process from Italy to France occurred during the ninetieth and twentieth centuries, it is of interest to see if French and Italian patients share common haplotypes for some of five mutations. For mutations p.Arg484Trp and p.Arg479X, completely different haplotypes are found in the two countries. The mutation p.Arg67Gln has been reported so far only in France and Italy.14 Two different haplotypes were found in French patients. One French patient shared a common allele for the intragenic D12S1677 marker with two unrelated Italian patients. This could be the indication of a common origin of the mutation but since the shared allele is rather frequent in the French population (allele 3 with frequency 0.225 in the French controls), it is more likely that the sharing only occurred by chance. For the p.Arg374Trp mutation, the common origin hypothesis is also possible with one Italian patient sharing alleles with three French patients at the two flanking markers D12S296 and D12S1677. These alleles, allele 5 and 1, respectively, are not present in the French control sample, and this might give some more weight to the hypothesis of a common origin of the mutation. For the p.Arg144X mutation, several patients from both countries shared the ?-6-144X-5-6-7 haplotype and a common origin is, thus, very likely and was hypothesized to compute the age of the MRCA with the ESTIAGE program. However, we cannot exclude that the p.Arg144X mutation had occurred twice on a common ancestral haplotype and that French and Italian patients had independent MRCA for this mutation. According to the present results, it is thus plausible that some of the ACVRL1 mutations had occurred in Italy and were brought to France several generations later by Italian migrants.
Age estimates of founder events
To estimate the age of the MRCA of the different mutations with founder effects, the method implemented in the ESTIAGE program requires the knowledge of the haplotypes associated with the mutation in the different patients. When only genotypes are available at the different markers, it is possible to reconstruct haplotypes using statistical methods such as the one implemented in the PHASE software and to consider the best haplotype reconstruction.21 Another possibility is to use a maximum parsimony principle as it was done here, where haplotypes were assigned to individuals to maximize the length of sharing around the mutation. This method appears to us intuitively more appropriate to the problem of the estimation of the age of a founder effect mutation than a method that will reconstruct haplotype statistically without accounting for the fact that patients have inherited an identical-by-descent mutation. It has also the advantage of being rather simple. We compared the results we obtained here with those obtained after reconstructing haplotypes with PHASE and found that they were in fact very similar with differences in age estimates of one or two generations (data not shown).
A second limitation of the method is the need to have marker allele frequency estimates from the population where patients are sampled. In this study, allele frequencies at the different markers were obtained from a sample of 20 French controls. A larger control sample could have been useful to obtain more precise estimates of allele frequencies, but this would probably not have led to important changes in our MRCA age estimates as the ESTIAGE method was shown to be robust to small misspecification in allele frequencies.15
A third and probably more serious limitation of the ESTIAGE method is the assumption of a star-shaped genealogy; that is, it is assumed that the different patients diverged independently from the ancestor. The impact of departure from the star-shaped genealogy assumption on age estimates depends on the population demography and is thus difficult to evaluate.22 In the case of the p.Arg144X mutation for which French and Italian patients were used together, it could be of interest to try to relax the star-shaped assumption by using a method such as one recently proposed by Hanein et al23 and that allow for bifurcating genealogies with two ancestors.
References
Plauchu H, de Chadarevian JP, Bideau A, Robert JM : Age-related clinical profile of hereditary hemorrhagic telangiectasia in an epidemiologically recruited population. Am J Med Genet 1989; 32: 291–297.
McAllister KA, Grogg KM, Johnson DW et al: Endoglin, a TGF-β binding protein of endothelial cells, is the gene for hereditary haemorrhagic telangiectasia type 1. Nat Genet 1994; 8: 345–351.
Johnson DW, Berg JN, Gallione CJ et al: A second locus for hereditary hemorrhagic telangiectasia maps to chromosome 12. Genome Res 1995; 5: 21–28.
Oh SP, Seki T, Goss KA et al: Activin receptor-like kinase 1 modulates transforming growth factor-β1 signaling in the regulation of angiogenesis. Proc Natl Acad Sci USA 2000; 97: 2626–2631.
Goumans MJ, Valdimarsdottir G, Itoh S, Rosendahl A, Sideras P, ten Dijke P : Balancing the activation state of the endothelium via two distinct TGF-β type I receptors. EMBO J 2002; 21: 1743–1753.
Gallione CJ, Repetto GM, Legius E et al: A combined syndrome of juvenile polyposis and hereditary hemorrhagic telangiectasia associated with mutations in MADH4 (SMAD4). Lancet 2004; 363: 852–859.
Cole SG, Begbie ME, Wallace GM, Shovlin CL : A new locus for hereditary haemorrhagic telangiectasia (HHT3) maps to chromosome 5. J Med Genet 2005; 42: 577–582.
Bayrak-Toydemir P, McDonald J, Akarsu N et al: A fourth locus for hereditary hemorrhagic telangiectasia maps to chromosome 7. Am J Med Genet A 2006; 140: 2155–2162.
Lesca G, Burnichon N, Raux G et al: Distribution of ENG and ACVRL1 (ALK1) mutations in French HHT patients. Hum Mutat 2006; 27: 598.
Lesca G, Plauchu H, Coulet F et al: Molecular screening of ALK1/ACVRL1 and ENG genes in hereditary hemorrhagic telangiectasia in France. Hum Mutat 2004; 23: 289–299.
Bideau A, Plauchu H, Brunet G, Robert J : Epidemiological investigation of Rendu-Osler disease in France: its geographical distribution and prevalence. Popul 1989; 44: 3–22.
Kjeldsen AD, Vase P, Green A : Hereditary haemorrhagic telangiectasia: a population-based study of prevalence and mortality in Danish patients. J Intern Med 1999; 245: 31–39.
Dakeishi M, Shioya T, Wada Y et al: Genetic epidemiology of hereditary haemorrhagic telangiectasia in a local community in the northern part of Japan. Hum Mutat 2002; 19: 140–148.
Olivieri C, Mira E, Delù G et al: Identification of 13 new mutations in the ACVRL1 gene in a group of 52 unselected Italian patients affected by hereditary haemorrhagic telangiectasia. J Med Genet 2002; 39: e39.
Génin E, Tullio-Pelet A, Begeot F, Lyonnet S, Abel L : Estimating the age of rare disease mutations: the example of Triple-A syndrome. J Med Genet 2004; 41: 445–449.
Bayrak-Toydemir P, McDonald J, Markewitz B et al: Genotype–phenotype correlation in hereditary hemorrhagic telangiectasia: mutations and manifestations. Am J Med Genet A 2006; 140: 463–470.
Cooper DN, Youssoufian H : The CpG dinucleotide and human genetic disease. Hum Genet 1988; 78: 151–155.
Heyer E, Brunet G : Généalogie et structure génétique de la population; in Bideau A, Brunet G (eds): Essai de démographie historique et de génétique des populations. Paris: Une population du Jura méridional du XVIIe siècle à nos jours, 2007, pp 159–171.
Brusgaard K, Kjeldsen AD, Poulsen L et al: Mutations in endoglin and in activin receptor-like kinase 1 among Danish patients with hereditary haemorrhagic telangiectasia. Clin Genet 2004; 66: 556–561.
Gallione CJ, Scheessele EA, Reinhardt D et al: Two common endoglin mutations in families with hereditary hemorrhagic telangiectasia in the Netherlands Antilles: evidence for a founder effect. Hum Genet 2000; 107: 40–44.
Stephens M, Smith NJ, Donnelly P : A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 2001; 68: 978–989.
Rosenberg NA, Hirsch AE : On the use of star-shapes genealogies in inference of coalescence times. Genetics 2003; 164: 1677–1682.
Hanein S, Perrault I, Gerber S et al: Population history and infrequent mutations: how old is a rare mutation? GUCY2D as a worked example. Eur J Hum Genet 2008; 16: 115–123.
Acknowledgements
We thank the patients and their colleagues from the French-Italian HHT Network who referred the patients. This work was supported by the following sources of funding: Fondation pour la Recherche Médicale, Rare Disease Network financed by INSERM and AFM (Association Française contre les Myopathies), Projet Hospitalier de Recherche Clinique 27-2004 (French Ministry of Health) and Fondazione Banca del Monte di Lombardia (Pavia, Italy).
Author information
Authors and Affiliations
Consortia
Corresponding author
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary information
Rights and permissions
About this article
Cite this article
Lesca, G., Genin, E., Blachier, C. et al. Hereditary hemorrhagic telangiectasia: evidence for regional founder effects of ACVRL1 mutations in French and Italian patients. Eur J Hum Genet 16, 742–749 (2008). https://doi.org/10.1038/ejhg.2008.3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2008.3
Keywords
This article is cited by
-
Sequence variations of ACVRL1 play a critical role in hepatic vascular malformations in hereditary hemorrhagic telangiectasia
Orphanet Journal of Rare Diseases (2020)
-
Future treatments for hereditary hemorrhagic telangiectasia
Orphanet Journal of Rare Diseases (2020)
-
The Stratified Population Screening of Hereditary Hemorrhagic Telangiectasia
Pathology & Oncology Research (2020)
-
Mutation affecting the proximal promoter of Endoglin as the origin of hereditary hemorrhagic telangiectasia type 1
BMC Medical Genetics (2017)
-
Increase of circulating endothelial cells in patients with Hereditary Hemorrhagic Telangiectasia
International Journal of Hematology (2015)