Abstract
To determine the phenotypic significance of copy number changes (CNCs) in the human genome, we performed genome-wide segmental aneuploidy profiling by BAC-based array-CGH of 278 unrelated patients with multiple congenital abnormalities and mental retardation (MCAMR) and in 48 unaffected family members. In 20 patients, we found de novo CNCs composed of multiple consecutive probes. Of the 125 probes making up these probably pathogenic CNCs, 14 were also found as single CNCs in other patients and 5 in healthy individuals. Thus, these CNCs are not by themselves pathogenic. Almost one out of five patients and almost one out of six healthy individuals in our study cohort carried a gain or a loss for any one of the recently discovered microdeletion/microduplication loci, whereas seven patients and one healthy individual showed losses or gains for at least two different loci. The pathogenic burden resulting from these CNCs may be limited as they were found with similar frequencies among patients and healthy individuals (P=0.165; Fischer's exact test), and several individuals showed CNCs at multiple loci. CNCs occurring specifically in our study cohort were enriched for components of the glutamate receptor family (GRIA2, GRIA4, GRIK2 and GRIK4) and genes encoding proteins involved in guiding cell localization during development (ATP1A2, GIRK3, GRIA2, KCNJ3, KCNJ10, KCNK17 and KCNK5). This indicates that disease cohort-specific compilations of CNCs may aid in identifying loci, genes and biological processes that contribute to the phenotype of patients.
Similar content being viewed by others
Introduction
With a prevalence of ∼2%, mental retardation is the most common reason for referral to medical genetic services.1 Unlike in patients with X-linked mental retardation,2 the genetic cause of mental retardation can rarely be pinpointed to a genetic locus or to a gene.3 Although the very genetic heterogeneity of mental retardation renders genetic linkage and association studies difficult, chromosomal aberrations, occurring with an incidence of 1 per 120 newborns,4 frequently provide a clue regarding the genetic locus underlying the phenotype of the affected child. Techniques allowing the detection of submicroscopic segmental aneuploidy have enabled us to pinpoint novel microdeletion and microduplication syndromes, such as CHARGE (coloboma, heart anomalies, chonal atresia, retardation, genital and ear anomalies),5 Peters Plus,6 recurrent 17q12 rearrangements,7 del(17)(q21),8, 9 and the 22q13.3 deletion,10 Pitt–Hopkins11 and thrombocytopenia-absent-radius syndromes12 (for reviews see Lee and Lupski3 and Slavotinek13). In addition, clinically inconsequential segmental deletions14, 15, 16 and segmental duplications in healthy individuals17 have been reported. Studies involving array-based comparative genome hybridization (array-CGH),18, 19 representational oligonucleotide microarray analysis,20 fosmid library insert mapping21 and SNP inheritance analyses22, 23, 24 indicate that copy number changes (CNCs) are frequently occurring polymorphisms in the genome of healthy individuals.25, 26, 27 The architecture of our chromosome complement, as it emerged during evolution, may be a driving force behind this type of genome diversity.26, 28, 29, 30, 31 Efforts to systematically compile CNCs have led to the identification of well over 6000 of such CNCs in DNA samples derived from lymphoblastoid cell lines of individuals of the HapMap population18 or from peripheral blood-derived DNA samples.32, 33, 34 The very existence of this type of structural genomic variability among apparently healthy individuals complicates a straightforward identification of pathogenetic segmental aneuploidy in patients.
To investigate whether CNCs may contribute to the complex spectrum of disorders consisting of combinations of mental retardation, developmental delay and multiple congenital anomalies (MCAMR), we determined the genomic locations and population frequencies of the CNCs shared among 278 patients and 48 of their unaffected family members seen in our institution. With this compilation of CNCs, we first determined the relative contribution of the microdeletion/microduplication syndromes that were recently discovered by array-based genome-wide segmental aneuploidy screening projects. Using gene prioritization tools (eg, Gene Annotation Tool to Help Explain Relationships (GATHER), Endeavour, Prioritizer), we secondly determined whether loci and genes pointing towards specific biochemical pathways were enriched in MCAMR patients by analyzing the 481 CNCs found exclusively in our cohort.
Materials and methods
Patient and reference populations
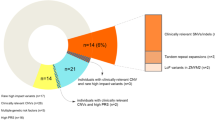
A total of 278 unrelated patients showing a combination of mental retardation and skeletal or facial dysmorphisms, according to the criteria described by De Vries et al35, and 48 of their unaffected parents were included in analyses of segmental aneuploidy by array-CGH. All patients were karyotyped at >550 band level. Patients with an abnormal karyotype, a subtelomeric anomaly or a known microdeletion or microduplication syndrome detected by fluorescence in situ hybridization (FISH) or Multiplex Ligation-mediated Probe Amplification were excluded. To investigate whether a CNC has occurred de novo in an affected child, the parents were also analyzed by array-CGH. Thus, 20 de novo CNCs composed of multiple consecutive probes, presumed to be pathogenic, were detected in 20 (7.2%) out of 278 patients.
For this study, we generated two reference pools by combining DNAs derived from peripheral blood samples of 50 unrelated and healthy male and of 50 unrelated and healthy female donors (age range: 22–63 years). The blood samples were obtained through the Department of Hematology of the UMC Utrecht. The amount of each contributing DNA sample was adjusted in such a manner that it comprised 2% of the total reference pool. In this way, CNCs occurring at a frequency larger than 2% in the general population are likely to be included in the reference pools.
Array-CGH
For array-CGH, 1 μg of sonicated genomic DNA from each patient (Tester) and a 1 μg sample of sonicated genomic DNA from the sex-mismatched reference pool were labelled with Cy3-dUTP and Cy5-dUTP (Amersham Biosciences, Little Chalfont, UK), respectively, using the BioPrime DNA Labeling System (Invitrogen, Carlsbad, CA, USA). These labelled DNAs were co-hybridized to the array-slides using a GeneTAC Hybstation (Genomic Solutions, Ann Arbor, MI, USA).5 The array has been created by spotting in triplicate DOP-PCR products of 3783 BAC DNA probes that cover the entire human genome with an average spacing of ∼0.7 Mb.5 Only BACs with a unique chromosomal location, as ascertained by FISH, have been included.36 After hybridization and washing, the slides were scanned and imaged on a ScanArray Express HT (Perkin Elmer, Wellesley, MA, USA) using ScanArray Express software (version 2.1). Data were analyzed with the GenePix Pro 5.0 software package (Axon Instruments, Union City, CA, USA), followed by LOWESS normalization using the Acuity 3.1 software package (Axon Instruments). After analysis, the averages of all triplicate spots were calculated. Subsequently, data were analyzed essentially according to Wong et al.32 Briefly, all spots with an aberrant shape, signal to noise ratios <5.0 or with an SD above 0.3 were excluded. Using the signals of all autosomal probes, the 95% confidence interval (95% CI) was calculated. All autosomal BACs outside the 95% CI with a P-value of <0.05 for being nonaberrant (two-sided Student's t-test) of all participants were tabulated in a single Excel worksheet. Using a confidence interval of 3.3 or even 4.0 SD gave smaller numbers of aberrant signals, but the same BACs were eventually identified (results not shown). Subsequently, the significantly aberrant signal populations of all participants thus obtained were compared with each other. As all precautions notwithstanding singly occurring aberrant signals could still represent false positives, these were excluded from further analyses. Thus, on an average, 1.96 aberrant signals per experiment were eliminated. This amounts to 0.052% of all BAC probes on the array. All aberrant signals occurring more than once were identified and their precise genomic locations were retrieved from the UCSC Genome Browser (Human Genome Build 36, March 2006). As we used a sex-mismatched reference sample, X and Y chromosome BACs are not informative for our analyses. Hence, our approach does not permit any inferences regarding CNCs of sex chromosomes. The array data analysis procedure is outlined in Figure 1.
Illumina Infinium HumanHap300 Genotyping BeadChip SNP array
For the Infinium HumanHap300 Genotyping BeadChip SNP array analysis, we used 750 ng of patient DNA and followed the protocol as described by the manufacturer (Illumina, San Diego, CA, USA).
Results
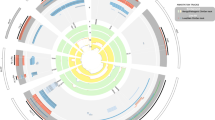
We performed genome-wide segmental aneuploidy profiling using a 3783 BAC-array with a functional genome coverage of 21.0% (for a definition of functional genome coverage see Coe et al37) of 278 unrelated MCAMR patients and 48 of their unaffected family members (see Materials and methods). In 326 hybridizations, we found a total of 12 486 significantly aberrant signals, which amounted to an average of 38.3 (SD of 19.5; range: 14–96) and a median of 34.0 aberrant signals per participant. After excluding false positives (see Materials and methods), we retained aberrant signals representing a total of 1308 distinct loci in our study cohort. As we compared DNA samples of a single proband with a reference composed of a mixture of 50 DNA samples (see Materials and methods), these loci represent directly observed recurrent CNCs. The genomic locations of all CNCs and their frequency of occurrence as losses and gains in our study population are shown in online Table 1. As an example, the data for chromosome 1 are displayed in Figure 2.
Recurrent CNCs specifically found in our MCAMR cohort. All CNCs mentioned in the DGV database have been omitted. Each dot represents a BAC probe according to its nucleotide position, following the March 2006 human reference sequence (NCBI Build 36.1). The ordinate position of each symbol represents the frequency of each CNC among MCAMR patients (closed symbol) and healthy individuals (open symbols). For each de novo loss and de novo gain found in our patients, red bars in the bottom decade and green bars in the top decade, respectively, have been added.
As part of our study, we detected CNCs involving multiple consecutive BAC probes in 20 out of 278 patients, which have arisen de novo and were therefore presumed to be pathogenic (Table 1)38. Several of these cases have been described in detail.39, 40, 41, 42, 43, 44 In addition, we found nine inherited segmental aneuploidies. The 20 de novo aberrations encompassed 125 BAC probes, 14 of which were also identified as single CNCs shared among at least 2 other patients and 5 among the 48 healthy individuals of our study population. Of these 125 BAC probes, 65 have been detected among healthy individuals and are listed in the Database of Genomic Variants (DGV, http://projects.tcag.ca/variation). This emphasizes that de novo segmental aneuploidies and recurrent single-probe CNCs represent two intrinsically linked sets of genomic loci and that CNCs at these loci are not per se pathogenic, although they may contribute to the patient's phenotype.
Out of the 3783 BACs on our array, 23 correspond to 11 of the microdeletion/microduplication syndromes that were recently discovered by genome-wide array-based screening efforts13, 45, 46 (Table 2). However, our array did not cover the 17q21.3 and the 16p11p12 microdeletion syndromes.8, 9 Thus, 23 BAC probes revealed 84 gains and losses (0.67% of all aberrant signals) in hybridizations of 76 DNA samples from 69 patients (21.6% of all patients) and 7 unaffected parents (14.8% of all parents). These frequencies among patients and healthy individuals are not significantly different (P=0.165; Fischer's exact test). Thus, close to one out of five individuals of our study cohort carried gains or losses at recurrent microdeletion/microduplication loci (Table 2). Hybridizations of seven patients and one healthy individual showed a loss or gain for two different microdeletion/microduplication loci. For five loci, at 1q21, 9q22.3, 15.q13.3, 16p13 and 20q13.13q13.2, frequencies of gains and losses in our study cohort exceeded 1% (Table 2). These data suggest that some of the recurrent microdeletion/microduplication loci are relative hotspots for segmental aneuploidy among patients with MCAMR and their parents. As CNCs at these recurrent microdeletion/microduplication loci occurred in healthy individuals, the pathogenic burden resulting from these CNCs may be limited, as has already been proposed by Ledbetter.47
Subsequently, we eliminated all CNCs that have previously been found as copy number variants in cohorts of healthy individuals (DGV; http://projects.tcag.ca/variation) and in those found among patients and healthy individuals in a study of amyotrophic lateral sclerosis.48 Thus, we retained 481 CNCs that were exclusively found among the 278 patients in our study cohort. Of these, 203 CNCs were shared at a >1% frequency among the patients. Subsequently, we determined whether the frequencies of losses and gains among these 203 retained CNCs were distributed according to a random generation and elimination model or were subjected to selective pressures. Out of these 203 CNCs, 30 (14.8%) occurred only as a loss and 31 (15.3%) only as a gain. This distribution of the 203 CNCs shared at >1% among the participants in our study cohort is not consistent with a random generation and elimination model, and thus indicates that these CNCs may have been subjected to selective pressures.
In an analogy to Wong et al,32 we assumed that CNCs are identical to the BAC probe representing them. This has been confirmed by SNP array mapping of break points in a small sample from our patient cohort (Table 1). On the basis of this assumption, we analyzed the gene content of the CNCs found in our study population. The 278 patients in our cohort shared 481 CNCs; 78 of these were found as a loss only and 89 as a gain only. No genes were covered by 26 of those CNCs occurring as a loss only and by 35 of those CNCs occurring as a gain only. All other CNCs were associated with annotated genes. For instance, RP11-96F24, which was not previously reported as a CNC, showed a gain in 22 patients, but not in any healthy individual of our study cohort, and no hemizygous loss. This BAC probe covers the glutathione S-transferase M3 (GSTM3) gene. Although an increased dosage for this gene is apparently well tolerated, haploinsufficiency may not be compatible with life.
To systematically determine whether the CNCs that were specifically found among the members of our MCAMR patient cohort covered genes that encoded shared cellular components, biological processes or molecular functions, we first determined to which Gene Ontologies (http://www.geneontology.org/GO.doc.shtml) these belong. To do so, we analyzed our data using GATHER developed by Chang and Nevins.49 Regardless of the frequencies of CNCs in our study cohort, each gene name was entered once. This allowed circumventing potential biases due to multiple listings of genes. We found that genes involved in potassium ion transport (GO:0006813) were specifically enriched for in the CNCs shared among members of our patient cohort with a P<0.0001 (χ2-test) and a Bayes factor of 9 (likelihood of association vs nonassociation). These genes were ABCC9, ATP1A2, ATP1A4, ATP1B2, GIRK3, GRIA2, GRIA4, GRIK2, GRIK4, KCNJ10, KCNJ3, KCNJ8, KCNK16, KCNK17, KCNK5, KCNK6, KCNN2, KCNN4, KCNV2, KCTD13, KCTD17, KCTD3 and SLC24A3. Of these 23 genes associated with potassium transport, seven (ATP1A2, GIRK3, GRIA2, KCNJ10, KCNJ3, KCNK17 and KCNK5) were also associated with the establishment of localization (GO:0051234). Thus, genes involved in guiding cell localization during development by controlling potassium transport seem to be selectively enriched among CNCs shared among MCAMR patients, and are not found in healthy individuals. Analysis of our CNC set using the Endeavour gene prioritization tool developed by Aerts et al50 and the Prioritizer tool developed by Franke et al,51 using different combinations of databases, corroborated this conclusion, although P-values remained slightly above a significance level of 0.05 (results not shown).
It should be noted that the BAC-array used in this study affords a relatively low-resolution mapping of the break points of CNCs. Thus, it is conceivable that assuming that the break points of CNCs coincide with the borders of BAC clones (in analogy to Wong et al32) may lead to a spurious inclusion of genes not involved in the observed CNC. This would lead to ‘noise’ in the gene prioritization analyses in such a way that no significance may be reached. In our analyses, and in those reported by Cuscó et al,52 still significant enrichment of certain Gene Ontologies has been found. Yet, in future studies, higher resolution platforms are recommended.
To determine whether some of these genes are part of one or several macromolecular networks, we used the Search Tool for Recurring Instances of Neighbouring Genes (STRING) developed by Snel et al,53 and found that genes from the glutamate receptor family (GRIA2, GRIA4, GRIK2 and GRIK4) form, on the basis of protein–protein interactions, a network with NSF, GRIP1, PRKCABP, GRIN1, GSR, RIPK1, PRKCG and GLUR6. In addition, the ATP-sensitive inward rectifier potassium channel genes KCNJ10, KCNJ3, KCNJ8 and GIRK3 may form heteromeric protein complexes with each other and with PSD95, KCNJ16, KCNJ5 and KCNJ6. These gene prioritization analyses indicate that the MCAMR-associated CNCs identified in our study may to some extent represent a distinct population of dosage-sensitive genes, and point towards shared biological processes in children with MCAMR.
Discussion
In this study, we addressed three questions. First, we determined whether the loci of de novo segmental aneuploidy in MCAMR patients are unique or overlap with the recurrent CNCs found among healthy relatives as well. Second, we tried to assess the pathogenic burden resulting from the microdeletion/microduplication syndromes recently discovered by genome-wide array-based segmental aneuploidy profiling efforts in our cohort of MCAMR patients. Third, we investigated whether the recurrent patient-specific CNCs in our study cohort harbour genes that may be involved in common biochemical pathways relevant to the phenotypes of MCAMR.
To do so, we compiled recurrent CNCs in a cohort of 278 unrelated MCAMR patients and 48 of their unaffected family members with a 3783 BAC-array. We used rigorous criteria to identify BAC probes showing aberrant signals without resorting to preset assumptions of a putative rate of aberration in each individual participant. In addition, similar to Wong et al,32we took several precautions to exclude false-positive signals. These attempts to obtain a ‘clean’ compilation of CNCs in our study cohort may have led to some false negatives. The patient cohort for our study was chosen such as to exclude aberrant karyotypes and subtelomeric anomalies or identifiable ‘classical’ microdeletion or microduplication syndromes (eg, the Smith–Magenis syndrome, the 22q11 syndrome). No additional selection criteria were applied. Therefore, our study cohort constitutes a reasonable representation of patients with multiple congenital anomalies and mental or motor retardation for which no diagnosis could be reached with standard diagnostic methods. To minimize potential population-based differences in haplotype compositions, we used a mixture of DNA samples from 50 healthy male or female volunteers. Finally, to exclude a spurious generation of CNCs by Epstein–Barr virus transformation,33, 54 we used DNA samples directly derived from freshly drawn peripheral blood samples only. In view of the precautions detailed above, the 1308 loci found are close to representing the true CNC content of our study cohort, as can be ascertained at the level of the functional genome coverage afforded by the array used (21%).37 As has been pointed out,25 a higher probe resolution and genome coverage may detect more CNCs, but the relevance of these smaller CNCs to the pathology in patients with multiple congenital anomalies and mental retardation remains unknown.
First, we determined whether the loci of de novo segmental aneuploidy in MCAMR patients are unique or overlap with the recurrent CNCs found among healthy relatives as well. Of the 125 BAC probes representing 20 de novo segmental aneuploidies among our 278 patients, 65 have been detected among healthy individuals in other studies (see DGV; http://projects.tcag.ca/variation) and 14 have also been identified as single CNCs shared among at least two other patients and five among the 48 healthy relatives of our study population. This emphasizes that de novo segmental aneuploidies and recurrent CNCs represent two intrinsically linked sets of genomic loci. In addition, these findings indicate that a mere CNC at any of these loci is neither per se pathogenic nor phenotypically neutral. However, hemizygosity for a certain gene, as provoked by a copy number loss, may ‘unmask’ an autosomal-recessive mutation in the retained allele.12, 43 CNCs showing a genomic gain, on the other hand, may contribute to the patient's phenotype if the genes contained therein encode proteins that form heteromeric complexes with other protein(s). Then, the balance of the participating protein components may be perturbed such that nonfunctional complexes with pathogenic consequences may be formed. This molecular mechanism may account for the apparent gene dosage sensitivity of certain genes, which are covered by these CNCs detected as both gains and losses in our study (eg, KCNJ10, KCNJ3 and GIRK3). Enrichment of this set of CNCs for these genes encoding components of the heteromeric ATP-sensitive inward rectifier potassium channels may represent such a case in point.
In addition, CNCs may affect the expression of genes in diploid chromosomal regions flanking the CNC.55, 56 Hence, CNCs may affect the phenotype by complex mechanisms in addition to and possibly different from altered gene dosage. Therefore, CNCs shared among unrelated patients with related phenotypes remain pathogenically relevant and merit further scrutiny. Thus, further investigations are needed to determine a potential contribution of a given CNC to the phenotype of an MCAMR patient.
Second, we tried to assess the relative pathogenic burden resulting from the recently reported novel microdeletion/microduplication syndromes.13, 45, 46 Only one out of the seven inherited and none of the de novo CNCs represented an example of these novel microdeletion/microduplication syndromes (case number 26 in Table 1), whereas all the other CNCs seem to be novel and unique. The latter has been a common finding in previous studies of cohorts of MCAMR patients (reviewed in Hochstenbach et al, 2009).57 Close to one out of five individuals in our study cohort carried gains or losses at recurrent microdeletion/microduplication loci. This indicates that the novel microdeletion/microduplication syndromes are frequently found among CNCs shared by our MCAMR patients and healthy individuals in our study cohort. As 76 out of 12 486 (0.61%) CNCs covered any of these 11 loci, the pathogenic burden of these CNCs may be relatively modest.47 As seven patients and one healthy individual showed a loss or gain for at least two different loci, these are not likely to be the sole cause of the patient's phenotype. In addition, patients with CNCs at some of these loci show rather divergent phenotypes.46, 58 Therefore, we infer that these loci may represent instances of ‘genomic unrest’, provoking a limited contribution to the aetiology of MCAMR and to the pathogenic burden in the population at large.
Third, our stepwise gene prioritization analyses indicate that the MCAMR-associated CNCs identified in our study point towards shared biological processes in children with MCAMR, including the glutamate receptor family (GRIA2, GRIA4, GRIK2 and GRIK4), which, by protein–protein interactions, forms a network with NSF, GRIP1, PRKCABP, GRIN1, GSR, RIPK1, PRKCG and GLUR6. In addition, the ATP-sensitive inward rectifier potassium channel genes, KCNJ10, KCNJ3, KCNJ8 and GIRK3, may form heteromeric protein complexes with each other and with PSD95, KCNJ16, KCNJ5 and KCNJ6. This suggests that the phenotypically diverse cohort of MCAMR patients may share a limited set of biochemical aetiologies.
In summary, our study of 278 patients with multiple congenital malformations and mental or motor retardation and 48 of their healthy relatives has unveiled widespread CNC. The CNCs occurring at a higher than 1% frequency in our study cohort seemed to occur as deletions only and as duplications only at less than expected rates. This indicates that the more frequently occurring CNCs have been subjected to selective pressure, suggesting that some of them may affect the phenotype. Therefore, these CNCs are not to be discarded, even if their contribution to the phenotype is not immediately obvious. Our finding that the set of CNCs shared among patients with MCAMR is enriched for genes encoding components of the ATP-sensitive inward rectifier potassium channel genes involved in guiding cell localization during development and members of the glutamate receptor family suggests that these biological processes may contribute to the phenotypes of patients with MCAMR. This hypothesis merits testing in replicating studies of other cohorts of patients and by using other biological pathway analysis algorithms. Eventually, patient–cohort-specific compilations of CNCs may serve as direct markers for genes that may help to identify loci, genes and biological processes contributing to the phenotype of patients.
References
Newman WG, Hamilton S, Ayres J et al: Array comparative genomic hybridization for diagnosis of developmental delay: an exploratory cost-consequences analysis. Clin Genet 2007; 71: 254–259.
Ropers HH : X-linked mental retardation: many genes for a complex disorder. Curr Opin Genet Dev 2006; 16: 260–269.
Lee JA, Lupski JR : Genomic rearrangements and gene copy-number alterations as a cause of nervous system disorders. Neuron 2006; 52: 103–121.
Nielsen J, Wohlert M : Chromosome abnormalities found among 34 910 newborn children: results from a 13-year incidence study in Arhus, Denmark. Hum Genet 1991; 87: 81–83.
Vissers LE, de Vries BB, Osoegawa K et al: Array-based comparative genomic hybridization for the genomewide detection of submicroscopic chromosomal abnormalities. Am J Hum Genet 2003; 73: 1261–1270.
Lesnik Oberstein SA, Kriek M, White SJ et al: Peters Plus syndrome is caused by mutations in B3GALTL, a putative glycosyltransferase. Am J Hum Genet 2006; 79: 562–566.
Mefford HC, Clauin S, Sharp AJ et al: Recurrent reciprocal genomic rearrangements of 17q12 are associated with renal disease, diabetes, and epilepsy. Am J Hum Genet 2007; 81: 1057–1069.
Koolen DA, Vissers LE, Pfundt R et al: A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat Genet 2006; 38: 999–1001.
Sharp AJ, Hansen S, Selzer RR et al: Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet 2006; 38: 1038–1042.
Cusmano-Ozog K, Manning MA, Hoyme HE : 22q13.3 deletion syndrome: a recognizable malformation syndrome associated with marked speech and language delay. Am J Med Genet C Semin Med Genet 2007; 145: 393–398.
Zweier C, Peippo MM, Hoyer J et al: Haploinsufficiency of TCF4 causes syndromal mental retardation with intermittent hyperventilation (Pitt-Hopkins syndrome). Am J Hum Genet 2007; 80: 994–1001.
Klopocki E, Schulze H, Strauss G et al: Complex inheritance pattern resembling autosomal recessive inheritance involving a microdeletion in thrombocytopenia-absent radius syndrome. Am J Hum Genet 2007; 80: 232–240.
Slavotinek AM : Novel microdeletion syndromes detected by chromosome microarrays. Hum Genet 2008; 124: 1–17.
Hand JL, Michels VV, Marinello MJ et al: Inherited interstitial deletion of chromosomes 5p and 16q without apparent phenotypic effect: further confirmation. Prenat Diagn 2000; 20: 144–148.
Sumption ND, Barber JC : A transmitted deletion of 2q13 to 2q14.1 causes no phenotypic abnormalities. J Med Genet 2001; 38: 125–127.
Barber JC : Directly transmitted unbalanced chromosome abnormalities and euchromatic variants. J Med Genet 2005; 42: 609–629.
Horvath JE, Bailey JA, Locke DP et al: Lessons from the human genome: transitions between euchromatin and heterochromatin. Hum Mol Genet 2001; 10: 2215–2223.
Iafrate AJ, Feuk L, Rivera MN et al: Detection of large-scale variation in the human genome. Nat Genet 2004; 36: 949–951.
Sharp AJ, Locke DP, McGrath SD et al: Segmental duplications and copy-number variation in the human genome. Am J Hum Genet 2005; 77: 78–88.
Sebat J, Lakshmi B, Troge J et al: Large-scale copy number polymorphism in the human genome. Science 2004; 305: 525–528.
Tuzun E, Sharp AJ, Bailey JA et al: Fine-scale structural variation of the human genome. Nat Genet 2005; 37: 727–732.
Conrad DF, Andrews TD, Carter NP et al: A high-resolution survey of deletion polymorphism in the human genome. Nat Genet 2006; 38: 75–81.
Hinds DA, Kloek AP, Jen M et al: Common deletions and SNPs are in linkage disequilibrium in the human genome. Nat Genet 2006; 38: 82–85.
McCarroll SA, Hadnott TN, Perry GH et al: Common deletion polymorphisms in the human genome. Nat Genet 2006; 38: 86–92.
Eichler EE : Widening the spectrum of human genetic variation. Nat Genet 2006; 38: 9–11.
Freeman JL, Perry GH, Feuk L et al: Copy number variation: new insights in genome diversity. Genome Res 2006; 16: 949–961.
McCarroll SA, Altshuler DM : Copy-number variation and association studies of human disease. Nat Genet 2007; 39: S37–S42.
Bailey JA, Gu Z, Clark RA et al: Recent segmental duplications in the human genome. Science 2002; 297: 1003–1007.
Stankiewicz P, Lupski JR : Genome architecture, rearrangements and genomic disorders. Trends Genet 2002; 18: 74–82.
Feuk L, Carson AR, Scherer SW : Structural variation in the human genome. Nat Rev Genet 2006; 7: 85–97.
Lupski JR : Genomic rearrangements and sporadic disease. Nat Genet 2007; 39: S43–S47.
Wong KK, deLeeuw RJ, Dosanjh NS et al: A comprehensive analysis of common copy-number variations in the human genome. Am J Hum Genet 2007; 80: 91–104.
Simon-Sanchez J, Scholz S, Fung HC et al: Genome-wide SNP assay reveals structural genomic variation, extended homozygosity and cell-line induced alterations in normal individuals. Hum Mol Genet 2007; 16: 1–14.
de Smith AJ, Tsalenko A, Sampas N et al: Array CGH analysis of copy number variation identifies 1284 new genes variant in healthy white males: implications for association studies of complex diseases. Hum Mol Genet 2007; 16: 2783–2794.
de Vries BB, White SM, Knight SJ et al: Clinical studies on submicroscopic subtelomeric rearrangements: a checklist. J Med Genet 2001; 38: 145–150.
Cheung VG, Nowak N, Jang W et al: Integration of cytogenetic landmarks into the draft sequence of the human genome. Nature 2001; 409: 953–958.
Coe BP, Ylstra B, Carvalho B et al: Resolving the resolution of array CGH. Genomics 2007; 89: 647–653.
Aston E, Whitby H, Maxwell T et al: Comparison of targeted and whole genome analysis of postnatal specimens using a commercially available array based comparative genomic hybridisation (aCGH) microarray platform. J Med Genet 2008; 45: 268–274.
de Pater JM, Nikkels PG, Poot M et al: Striking facial dysmorphisms and restricted thymic development in a fetus with a 6-megabase deletion of chromosome 14q. Pediatr Dev Pathol 2005; 8: 497–503.
de Pater JM, Poot M, Beemer FA et al: Virilization of the external genitalia and severe mental retardation in a girl with an unbalanced translocation 1;18. Eur J Med Genet 2006; 49: 19–27.
Poot M, Kroes HY, V D Wijst SE et al: Dandy-Walker complex in a boy with a 5 Mb deletion of region 1q44 due to a paternal t(1;20)(q44;q13.33). Am J Med Genet 2007; 143A: 1038–1044.
Poot M, Eleveld MJ, van 't Slot R et al: Proportional growth failure oculocutaneous albinism in a girl with a 6 87 Mb deletion of region 15q26.2 → qter. Eur J Med Genet 2007; 50: 432–440.
Flipsen-ten Berg K, van Hasselt PM, Eleveld MJ et al: Unmasking of a hemizygous WFS1 gene mutation by a chromosome 4p deletion of 8.3 Mb in a patient with Wolf-Hirschhorn syndrome. Eur J Hum Genet 2007; 15: 1132–1138.
Özgen HM, Staal WG, Barber JC et al: A novel 6.14 Mb duplication of chromosome 8p21 in a patient with autism and self mutilation. J Autism Dev Disord 2009; 39: 322–329.
Sharp AJ, Mefford HC, Li K et al: A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet 2008; 40: 322–328.
Mefford HC, Sharp AJ, Baker C et al: Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med 2008; 359: 1685–1699.
Ledbetter DH : Cytogenetic technology—genotype and phenotype. N Engl J Med 2008; 359: 1728–1730.
Blauw HM, Veldink JH, van Es MA et al: Copy-number variation in sporadic amyotrophic lateral sclerosis: a genome-wide screen. Lancet Neurol 2008; 7: 319–326.
Chang JT, Nevins JR : GATHER: a systems approach to interpreting genomic signatures. Bioinformatics 2006; 22: 2926–2933.
Aerts S, Lambrechts D, Maity S et al: Gene prioritization through genomic data fusion. Nat Biotechnol 2006; 24: 537–544.
Franke L, van Bakel H, Fokkens L et al (2006) Reconstruction of a functional human gene network, with an application for prioritizing positional candidate genes. Am J Hum Genet 78: 1011–1025.
Cuscó I, Medrano A, Gener B et al: Autism-specific copy number variants further implicate the phosphatidylinositol signaling pathway and the glutamatergic synapse in the etiology of the disorder. Hum Mol Genet 2009; 18: 1795–1804.
Snel B, Lehmann G, Bork P et al: STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res 2000; 28: 3442–3444.
Redon R, Ishikawa S, Fitch KR et al: Global variation in copy number in the human genome. Nature 2006; 444: 444–454.
Merla G, Howald C, Henrichsen CN et al: Submicroscopic deletion in patients with Williams-Beuren syndrome influences expression levels of the nonhemizygous flanking genes. Am J Hum Genet 2006; 79: 332–341.
Reymond A, Henrichsen CN, Harewood L et al: Side effects of genome structural changes. Curr Opin Genet Dev 2007; 17: 381–386.
Hochstenbach R, van Binsbergen E, Engelen J et al: Array analysis and karyotyping: workflow consequences based on a retrospective study of 36 325 patients with idiopathic developmental delay in the Netherlands. Eur J Med Genet 2009; e-pub ahead of print 9 April 2009.
Koolen DA, Sharp AJ, Hurst JA et al: Clinical and molecular delineation of the 17q21.31 microdeletion syndrome. J Med Genet 2008; 45: 710–720.
Acknowledgements
We kindly acknowledge the help of Hylke M Blauw, Jan H Veldink and Leonard H van den Berg (Department of Neurology, University Medical Centre Utrecht) with the processing of the data set.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary information
Rights and permissions
About this article
Cite this article
Poot, M., Eleveld, M., van 't Slot, R. et al. Recurrent copy number changes in mentally retarded children harbour genes involved in cellular localization and the glutamate receptor complex. Eur J Hum Genet 18, 39–46 (2010). https://doi.org/10.1038/ejhg.2009.120
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2009.120