Abstract
Adiponectin is an adipocyte-derived protein that is down-regulated in obesity-linked disorders. Variants of the adiponectin gene (ADIPOQ) have been shown to affect adiponectin level. We have now examined the relation of polymorphisms of ADIPOQ to adiponectin concentration and to metabolic disorders in the Kita-Nagoya Genomic Epidemiology study, a population-based study of elderly Japanese. The genomic region including ADIPOQ was genotyped for 30 single nucleotide polymorphisms in 500 subjects of a screening population with the use of a fluorescence- or colorimetry-based allele-specific DNA primer–probe assay system. Four polymorphisms were then selected for genotyping in an additional 2797 subjects. Serum adiponectin level was negatively associated with metabolic abnormalities after adjustment for age and sex. The minor alleles of the rs1656930, Ile164Thr, and rs9882205 polymorphisms were associated with a low serum adiponectin level. Whereas the minor alleles of rs1656930 and rs9882205 were common (minor allele frequency of 6.2 and 38.5%, respectively), that of Ile164Thr was rare (0.9%). The minor allele of rs1656930 was positively associated with systolic blood pressure and the prevalence of hypertension. The association of rs1656930 with adiponectin level was replicated in an independent population. A subject with the 164Thr/Thr genotype had an extremely low serum adiponectin level (0.6 μg/ml) and the phenotype of metabolic syndrome. Our results suggest that a common variant of ADIPOQ, the minor allele of rs1656930, is associated with hypoadiponectinemia and hypertension. Screening for a common genetic background underlying low adiponectin levels might provide important information for assessment and management of metabolic disorders.
Similar content being viewed by others
Introduction
Metabolic syndrome (MetS) is characterized by a cluster of insulin resistance, obesity, dyslipidemia, and hypertension, and is a common cause of cardiovascular disease.1, 2 The prevalence of MetS in Japan has increased over recent decades as a result of changes in diet and physical activity.3 In addition to its role in energy storage, adipose tissue functions as an endocrine organ by secreting various bioactive substances or adipocytokines. These molecules are able to directly or indirectly affect the pathogenesis of metabolic disorders.4, 5
Adiponectin is an adipocytokine whose concentration is reduced in obesity.6 Low plasma adiponectin levels (hypoadiponectinemia) are also associated with type II diabetes,7, 8, 9, 10 with high and low circulating concentrations of triglyceride and high-density lipoprotein (HDL) cholesterol,11 respectively, with hypertension,12 and with coronary artery disease.13, 14, 15 These observations suggest that adiponectin might be a useful biomarker for management of metabolic disorders. Evidence also indicates that adiponectin has beneficial effects on insulin resistance,16 endothelial function, hypertension,17, 18, 19 ischemic heart disease, and atherosclerosis.20, 21 Adiponectin may, therefore, be a key determinant of the development of metabolic disorders.
Adiponectin is encoded by the gene ADIPOQ, which has been mapped to chromosome 3q27.22 Variants of ADIPOQ have been associated with adiponectin level and metabolic traits.23 Case–control studies showed that the Ile164Thr polymorphism of ADIPOQ was associated with hypoadiponectinemia, type II diabetes, hypertension, and coronary artery disease in the Japanese.12, 24, 25 Other such studies found that the rs266729 (–11377C → G) polymorphism of ADIPOQ was associated with type II diabetes in Caucasians.26, 27 The rs1501299 (+276G → T) polymorphism was also shown to be associated with adiponectin level and type II diabetes in Caucasians as well as in obese Japanese.28, 29 However, the clinical relevance of ADIPOQ variants to metabolic traits has remained to be fully explored in a population-based cross-sectional study of the Japanese population. We have now examined the relation of ADIPOQ variants to serum adiponectin level and metabolic traits in such a study. Our data indicate that a common variant of ADIPOQ, the minor allele of rs1656930, was associated with low adiponectin levels and hypertension. The association of this polymorphism with hypoadiponectinemia was replicated in an independent population.
Subjects and methods
Study subjects
The Kita-Nagoya Genomic Epidemiology (KING) study (ClinicalTrials.gov identifier: NCT00262691) is a population-based cross-sectional study based in Kita-Nagoya, Japan, for which a total of 3975 volunteers were recruited from participants of annual health checkups and which has been described in detail previously.30 Of these volunteers, 3297 individuals (1456 men, 1841 women; age, 63.6±6.5 years) whose serum adiponectin concentration was measured were enrolled in this study. A replication study included 750 men (57.8±3.1 years) recruited from Japanese company employee volunteers who were considered to be engaged mostly in light labor (described in detail under ClinicalTrials.gov identifier NCT00408824). A physical examination was performed and a medical history was obtained for each subject at an annual health checkup. Body mass index (BMI), waist circumference (at the midpoint between the lowest rib and the iliac crest after normal expiration), and blood pressure were measured for all subjects. The study protocol was approved by the ethics committees of Aichi-Gakuin University School of Dentistry and Nagoya University School of Medicine, and all participants provided written informed consent.
Definitions of risk factors
The overweight condition was defined as a BMI of ≥25 kg/m.2 Hypertension was defined as systolic blood pressure of ≥130 mm Hg or diastolic blood pressure of ≥85 mm Hg on repeated measurement, or as treatment with antihypertensive medication. Hyperglycemia was defined as a fasting serum glucose concentration of ≥100 mg/dl or a hemoglobin (Hb) A1c content of ≥5.9%, or as treatment with antidiabetes agents. Dyslipidemia was defined as a serum concentration of total cholesterol of ≥220 mg/dl, a serum triglyceride concentration of ≥150 mg/dl, or a serum HDL-cholesterol concentration of <40 mg/dl in men or <50 mg/dl in women, or as treatment for dyslipidemia. MetS was defined according to the recent International Diabetes Federation consensus criteria,1 which stipulate central obesity as being essential for the diagnosis.
Blood collection
Blood samples were obtained from subjects in the fasted condition for measurement of serum biomarkers and extraction of DNA. After the subject had rested for 10 min in the sitting position, vital signs were recorded and 14 ml of blood were collected from the antecubital vein into tubes containing EDTA. The blood was centrifuged at 3000 rpm for 15 min at 4°C, and the resulting supernatant was stored at −80°C until analysis. Serum adiponectin level was determined with the use of a latex turbidometric immunoassay (Otsuka Pharmaceutical Corporation, Osaka, Japan). Glucose, HbA1c, and lipids were measured by standard methods.
Genotyping of polymorphisms
Genomic DNA was isolated with the use of a FlexGene DNA kit (Qiagen, Chatsworth, CA, USA). Genotyping of 30 single nucleotide polymorphisms (SNPs) of ADIPOQ was performed with a fluorescence- or colorimetry-based allele-specific DNA primer–probe assay system (Toyobo Gene Analysis, Tsuruga, Japan) as described previously.31 To confirm the accuracy of genotyping, we randomly selected 100 DNA samples and subjected them to polymerase chain reaction (PCR) and restriction fragment length polymorphism analysis or to direct DNA sequencing of the PCR products. In each instance, the genotype determined by the allele-specific DNA primer–probe assay system was identical to that determined by the confirmatory methods. Furthermore, we genotyped all samples with a positive control (90 samples with a positive control at a time), and we confirmed that the positive control was genotyped correctly. The genotyping success rate was >99%, and all the SNPs were found to be in Hardy–Weinberg equilibrium (P>0.05 for all). Primer and probe sequences as well as PCR conditions for genotyping are shown in Supplementary Table 1.
Selection of SNPs
In an initial screening study, we genotyped 30 SNPs spanning the ADIPOQ locus in 500 subjects randomly selected from the total study population. The relation of these SNPs to serum adiponectin level was assessed, and those that showed such an association were genotyped in the remaining 2797 subjects. The 30 SNPs examined encompassed the genomic region (25 kbp) containing ADIPOQ exons and introns plus 8 kbp upstream and 1 kbp downstream (chromosome 3: genome positions 188034376–188059821) and were selected from those listed in the National Center for Biotechnology Information database dbSNP build 129 or those described in published studies. Several of the SNPs have previously been associated with adiponectin level32 and metabolic disorders including obesity,33, 34 type II diabetes mellitus,24, 28 or cardiovascular disease.25 In addition, we only selected SNPs for which the minor allele frequency (MAF) was shown to be >1% in Japanese subjects. The average distance between analyzed SNPs was 0.85 kb.
Statistical analysis
Allele frequencies were calculated by allele counting. The χ2 test was used to identify departures from Hardy–Weinberg equilibrium. The relation between serum adiponectin level and various parameters or ADIPOQ variants was evaluated by simple linear regression analysis or multivariate regression analysis. The relation of ADIPOQ genotype to continuous clinical parameters was evaluated by analysis of variance or analysis of covariance, with Scheffe's test used for post hoc analysis. Categorical variables were compared among genotypes with the use of the χ2 test. Student's t-test was used for dominant model analysis. Statistical analysis was performed with SPSS version 14.0 software (SPSS, Chicago, IL, USA). For continuous variables, results are presented as means±SD. Given that the distributions of serum adiponectin and triglyceride concentrations were skewed, values were logarithmically transformed for statistical analysis. In the screening study, the cutoff P-value was defined as <0.1 for simple linear regression analysis in order to avoid false negative results. Otherwise, a P-value of <0.05 was considered statistically significant. Haplotype analysis to detect linkage disequilibrium (LD) was performed with Haploview 4.0 software (Broad Institute, Cambridge, MA, USA).
Results
Clinical characteristics
The clinical characteristics of the KING study and replication study participants are shown in Table 1. The 3297 subjects of the KING study had a mean age of 63.6±6.5 years, with 55.8% of them being female. The average serum adiponectin level was 10.9 μg/ml (95% confidence interval (CI), 10.7–11.1 μg/ml), with that in women being significantly higher than that in men (Table 1; Supplementary Table 2). The 750 male subjects of the replication study had a mean age of 57.8±3.1 years and an average serum adiponectin level of 8.5 μg/ml (95% CI, 8.2–8.8 μg/ml).
Relation of adiponectin level to clinical parameters
We first examined the relation between serum adiponectin concentration and clinical parameters in all KING study subjects (Supplementary Table 2). Serum adiponectin level was positively associated with serum HDL-cholesterol concentration and negatively associated with BMI, blood pressure, serum concentrations of low-density lipoprotein cholesterol and triglyceride, fasting serum glucose concentration, and HbA1c level by simple linear regression analysis and by multivariate regression analysis with adjustment for age and sex. Consistent with previous observations,11 it also showed a significant association with hypertension, hyperglycemia, dyslipidemia, and the overweight condition.
Selection of SNPs in ADIPOQ and analysis of LD
We performed a screening study for 30 polymorphisms in and around ADIPOQ with 500 subjects randomly selected from the 3297 participants of the KING study (Table 1; Figure 1). Consistent with previous observations,32 we detected two LD blocks, and several SNPs also showed strong pairwise LD (Figure 2). SNPs rs4632532 (–19148C → T), rs1648707 (–19119C → A), and rs822388 (–8564T → G) were in strong LD (r2>0.95), as were rs16861194 (−11426A → G) and rs60806105 (–11156del/insCA); rs822391 (–7045T → C), rs822394 (–4120C → A), and rs822396 −3971A → G); rs2241766 (+45T → G), rs2241767 (+349A → G), and rs2082940 (+3317T → C); and rs3774261 (+712G → A), +2019del/insA, and rs1063538 (+3336T → C). Minor alleles of four SNPs (rs17300539 (−11391G → A), Gly90Ser, rs17366743 (Tyr111His), and Arg112Cys) were not found (MAF=0). Among the 30 SNPs analyzed, rs1656930 (−17991G → A), rs9882205 (–450A → G), and Ile164Thr were significantly associated with serum adiponectin level (P=0.034, 0.054, and 9.0 × 10–4, respectively) (Table 2).
Schematic representation of the genomic structure and polymorphic variants of ADIPOQ. Open and closed boxes represent noncoding and coding regions of exons, respectively. Nucleotide positions are relative to the translation initiation codon (ATG). Asterisks indicate rare polymorphisms that result in amino-acid substitutions.
Relation of ADIPOQ SNPs to serum adiponectin concentration and metabolic traits
The three SNPs associated with serum adiponectin level in the screening study as well as rs266729 (−11377C → G), the minor allele of which was previously associated with hypoadiponectinemia and type II diabetes,26 were genotyped in the full complement of 3297 subjects and examined for their relation to serum adiponectin concentration. The minor alleles of rs1656930, rs9882205, and Ile164Thr, but not that of rs266729, were associated with low serum adiponectin levels by simple linear regression analysis. These associations remained significant in multiple regression analysis after adjustment for age and sex either alone or together with other clinical parameters including BMI (Table 3). The minor allele of rs1656930, which was strongly associated with hypoadiponectinemia (Figure 3), was relatively common (MAF=6.2%). In contrast, the minor allele of Ile164Thr, which showed the strongest association with low serum adiponectin levels, was rare (MAF=0.9%). A 70-year-old man homozygous for the minor allele of Ile164Thr had the lowest adiponectin level (0.6 μg/ml) among the study population and was diagnosed with MetS, including hypertension and dyslipidemia. The minor allele of rs9882205 was common (MAF=38.5%) and showed a weak but significant association with hypoadiponectinemia. Conditional regression analysis with adjustment for each SNP indicated that rs1656930, Ile164Thr, and rs9882205 were independently associated with serum adiponectin level, although Ile164Thr and rs9882205 showed evidence of possible interdependence (Supplementary Table 3).
A replication study confirmed the association of the minor alleles of rs1656930 and Ile164Thr with serum adiponectin level in an independent population, although this population did not include any subjects who were homozygous for the minor alleles of these SNPs (Table 3). No significant association of rs9882205 and serum adiponectin level was detected in the replication study.
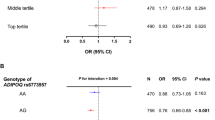
Finally, we assessed the relation of the four genotyped SNPs to clinical parameters in the KING study participants. The minor allele of rs1656930 was positively associated with systolic blood pressure and the prevalence of hypertension (Table 4). Indeed, all subjects homozygous for the minor allele of rs1656930 had hypertension. This association with systolic blood pressure remained significant in multiple regression analysis after adjustment for adiponectin level (Supplementary Table 4). The minor allele of Ile164Thr was associated with the prevalence of MetS (P<0.03), but not with that of the other metabolic disorders including hypertension. The minor alleles of rs9882205 and rs266729 were not associated with any clinical parameters (data not shown).
Discussion
We have performed a fine-mapping analysis of ADIPOQ and assessed the relation of gene variants to serum adiponectin concentration in a population-based study. As far as we are aware, this study provides the first demonstration of an association between a common variant of ADIPOQ (the minor allele of rs1656930) and hypoadiponectinemia.
ADIPOQ SNPs and adiponectin level
The SNP rs1656930 (−17991G → A) is located 6.6 kbp upstream of the transcription start site of ADIPOQ. The adiponectin gene contains two regions of high LD, one encompassing the promoter region and the other including intron 2 as well as the coding and 3′ untranslated regions of exon 3.32 We confirmed that the LD block in the promoter region extended to and included rs1656930 (Figure 2). It is thus possible that rs1656930 affects transcriptional activity or is in high LD with an unknown SNP in this region that affects such activity. The rs1656930 and rs864265 SNPs are in perfect pairwise LD (r2=1) in the HapMap database, but we did not genotype this latter SNP in this study. The combined group of subjects with the GA or AA genotype of rs1656930 had a significantly lower adiponectin level than did those with the GG genotype. This difference was also apparent when men and women were analyzed separately (Figure 3).
We found that the minor allele of the Ile164Thr polymorphism of ADIPOQ was also significantly associated with low adiponectin levels, consistent with previous observations.12, 25 Case–control studies have shown that heterozygosity for the Ile164Thr polymorphism was associated with metabolic abnormalities including hypertension and type II diabetes in Japanese.12, 24 We have now found that this polymorphism was associated with the prevalence of MetS, but not with that of hypertension or hyperglycemia. One subject homozygous for the Thr allele of this polymorphism had a markedly low adiponectin level (0.6 μg/ml) and features of MetS. As far as we are aware, a subject with the Thr/Thr genotype for this SNP has not been previously described. The discrepancy between our current data and the previous results may be due to differences in age or the frequency of hypertension or diabetes among study populations.
Whereas rs1501299 (+276G → T) was previously found to be associated with adiponectin level in obese Japanese subjects,28 it was not associated with serum adiponectin concentration in our initial screening study. In the Framingham Offspring study,35 two SNPs in the promoter region of ADIPOQ (rs822387, rs17300539) were shown to be strongly associated with plasma adiponectin level, whereas rs17366743 (Tyr111His) was associated with the prevalence of type II diabetes. However, in this study, none of these three SNPs was associated with adiponectin level or hyperglycemia in the screening population.
A genome-wide association study recently showed that rs17366568 and eight other SNPs within ADIPOQ were associated with plasma adiponectin level in Europeans.36 Although we did not genotype rs17366568, rs1656930 is independent of this SNP (r2=0) in the HapMap database. Of the other eight SNPs, only rs822394 showed possible pairwise LD (r2=0.585) with rs1656930 in the HapMap database. However, in this study, unlike rs1656930, rs822394 did not have a significant effect on serum adiponectin level (Table 2). Moreover, rs864265, which is in perfect LD with rs1656930, showed no significant association with adiponectin level in the European study.
The discrepancies in findings among these various studies may result from underlying differences in the study populations including the frequencies of ADIPOQ polymorphisms.
ADIPOQ SNPs and metabolic traits
ADIPOQ has been studied previously as a candidate gene for metabolic disorders,22 and the relation of gene variants to circulating adiponectin levels has long been recognized.23, 37 Despite the strong association between adiponectin concentration and metabolic disorders described in various studies, other analyses have failed to detect a significant association between ADIPOQ variants and metabolic phenotype.36, 38 We also detected few associations between metabolic abnormalities and the selected variants of ADIPOQ that are present within genomic regions previously shown to be associated with metabolic phenotype. We had a power of 100% in the KING study to detect associations of ADIPOQ variants with metabolic abnormalities similar to those described by Iwashima et al12 (assuming α=0.05), but the previously observed association of Ile164Thr with hypertension was not replicated in our study. Such discrepancies may reflect the multiple genetic or environmental factors that contribute to metabolic disorders.
Adiponectin and hypertension
We found that only the minor allele of rs1656930 was positively associated with systolic blood pressure and the prevalence of hypertension. Of note, the increase in systolic blood pressure associated with the minor allele of this SNP was apparent only for homozygotes (Table 4). Although this allele was also associated with a low adiponectin level, the association with systolic blood pressure remained significant after adjustment for adiponectin level. Hypertension is a complex disorder that results from the interplay of genetic and environmental influences. Hypoadiponectinemia has been implicated as an important risk factor for hypertension.12, 39 Hypertensive individuals with insulin resistance were also found to have a low adiponectin level.18, 40 Plasma adiponectin concentrations were significantly lower in men with hypertension than in normotensive men and were negatively correlated with blood pressure in subjects without hypertension.12 In addition, adiponectin levels were negatively correlated with blood pressure in normotensive subjects without abnormal glucose tolerance.12 Consistent with these clinical observations, adiponectin knockout mice manifest impaired endothelium-dependent vasodilation on an atherogenic diet17 as well as salt-sensitive hypertension.41 Adiponectin supplementation ameliorated elevated blood pressure in both obese KKAy mice and salt-fed adiponectin knockout mice.41 These results suggest that hypoadiponectinemia contributes to the development of hypertension.
Study limitations
In this study, there was no subject homozygous for the minor allele of rs1656930 in the replication population, reflecting the relatively small number of subjects in this population (n=750) compared with the total number of subjects in the KING study (n=3297). We were thus not able to confirm the association of the minor allele of rs1656930 with hypertension in the replication study. We had a power of 22.6% in the replication study to detect effects similar to those observed in the KING study (assuming α=0.05). Verification that rs1656930 is associated with hypertension will thus require independent confirmation in adequately powered samples. Similarly, we could not confirm the association of rs9882205 with serum adiponectin level in the replication study.
Conclusion
Our data demonstrate an association of the minor allele of the rs1656930 polymorphism of ADIPOQ both with a low serum adiponectin level and with the prevalence of hypertension in the Japanese population. Hypoadiponectinemia resulting from physical inactivity and excessive calorie intake is thought to contribute to the development of metabolic disorders and cardiovascular disease. Screening for a common genetic background of hypoadiponectinemia may, therefore, provide important information for the assessment and management of metabolic disorders.
References
Alberti KGMM, Zimmet P, Shaw J : Metabolic syndrome—a new world-wide definition. A Consensus Statement from the International Diabetes Federation. Diabet Med 2006; 23: 469–480.
Lakka H-M, Laaksonen DE, Lakka TA et al: The metabolic syndrome and total and cardiovascular disease mortality in middle-aged men. JAMA 2002; 288: 2709–2716.
Yoshiike N, Seino F, Tajima S et al: Twenty-year changes in the prevalence of overweight in Japanese adults: the National Nutrition Survey 1976-95. Obes Rev 2002; 3: 183–190.
Matsuzawa Y, Funahashi T, Kihara S, Shimomura I : Adiponectin and metabolic syndrome. Arterioscler Thromb Vasc Biol 2004; 24: 29–33.
Vettor R, Milan G, Rossato M, Federspil G : Review article: adipocytokines and insulin resistance. Aliment Pharmacol Ther 2005; 22: 3–10.
Weyer C, Funahashi T, Tanaka S et al: Hypoadiponectinemia in obesity and type 2 diabetes: close association with insulin resistance and hyperinsulinemia. J Clin Endocrinol Metab 2001; 86: 1930–1935.
Hotta K, Funahashi T, Arita Y et al: Plasma concentrations of a novel, adipose-specific protein, adiponectin, in type 2 diabetic patients. Arterioscler Thromb Vasc Biol 2000; 20: 1595–1599.
Lindsay RS, Funahashi T, Hanson RL et al: Adiponectin and development of type 2 diabetes in the Pima Indian population. Lancet 2002; 360: 57–58.
Spranger J, Kroke A, Möhlig M et al: Adiponectin and protection against type 2 diabetes mellitus. Lancet 2003; 361: 226–228.
Li S, Shin HJ, Ding EL, van Dam RM : Adiponectin levels and risk of type 2 diabetes: a systematic review and meta-analysis. JAMA 2009; 302: 179–188.
Ryo M, Nakamura T, Kihara S et al: Adiponectin as a biomarker of the metabolic syndrome. Circ J 2004; 68: 975–981.
Iwashima Y, Katsuya T, Ishikawa K et al: Hypoadiponectinemia is an independent risk factor for hypertension. Hypertension 2004; 43: 1318–1323.
Ouchi N, Kihara S, Arita Y et al: Novel modulator for endothelial adhesion molecules: adipocyte-derived plasma protein adiponectin. Circulation 1999; 100: 2473–2476.
Kumada M, Kihara S, Sumitsuji S et al: Association of hypoadiponectinemia with coronary artery disease in men. Arterioscler Thromb Vasc Biol 2003; 23: 85–89.
Otsuka F, Sugiyama S, Kojima S et al: Hypoadiponectinemia is associated with impaired glucose tolerance and coronary artery disease in non-diabetic men. Circ J 2007; 71: 1703–1709.
Snijder MB, Heine RJ, Seidell JC et al: Associations of adiponectin levels with incident impaired glucose metabolism and type 2 diabetes in older men and women: the Hoorn Study. Diabetes Care 2006; 29: 2498–2503.
Ouchi N, Ohishi M, Kihara S et al: Association of hypoadiponectinemia with impaired vasoreactivity. Hypertension 2003; 42: 231–234.
Francischetti EA, Celoria BMJ, Duarte SFP et al: Hypoadiponectinemia is associated with blood pressure increase in obese insulin-resistant individuals. Metabolism 2007; 56: 1464–1469.
Huang K-C, Chen C-L, Chuang L-M, Ho S-R, Tai T-Y, Yang W-S : Plasma adiponectin levels and blood pressures in nondiabetic adolescent females. J Clin Endocrinol Metab 2003; 88: 4130–4134.
Iwashima Y, Horio T, Suzuki Y et al: Adiponectin and inflammatory markers in peripheral arterial occlusive disease. Atherosclerosis 2006; 188: 384–390.
Pischon T, Girman CJ, Hotamisligil GS, Rifai N, Hu FB, Rimm EB : Plasma adiponectin levels and risk of myocardial infarction in men. JAMA 2004; 291: 1730–1737.
Hu E, Liang P, Spiegelman BM : AdipoQ is a novel adipose-specific gene dysregulated in obesity. J Biol Chem 1996; 271: 10697–10703.
Gable DR, Hurel SJ, Humphries SE : Adiponectin and its gene variants as risk factors for insulin resistance, the metabolic syndrome and cardiovascular disease. Atherosclerosis 2006; 188: 231–244.
Kondo H, Shimomura I, Matsukawa Y et al: Association of adiponectin mutation with type 2 diabetes : a candidate gene for the insulin resistance syndrome. Diabetes 2002; 51: 2325–2328.
Ohashi K, Ouchi N, Kihara S et al: Adiponectin I164T mutation is associated with the metabolic syndrome and coronary artery disease. J Am Coll Cardiol 2004; 43: 1195–1200.
Vasseur F, Helbecque N, Dina C et al: Single-nucleotide polymorphism haplotypes in the both proximal promoter and exon 3 of the APM1 gene modulate adipocyte-secreted adiponectin hormone levels and contribute to the genetic risk for type 2 diabetes in French Caucasians. Hum Mol Genet 2002; 11: 2607–2614.
Schwarz PEH, Towers GW, Fischer S et al: Hypoadiponectinemia is associated with progression toward type 2 diabetes and genetic variation in the ADIPOQ gene promoter. Diabetes Care 2006; 29: 1645–1650.
Hara K, Boutin P, Mori Y et al: Genetic variation in the gene encoding adiponectin is associated with an increased risk of type 2 diabetes in the Japanese population. Diabetes 2002; 51: 536–540.
Filippi E, Sentinelli F, Trischitta V et al: Association of the human adiponectin gene and insulin resistance. Eur J Hum Genet 2003; 12: 199–205.
Asano H, Izawa H, Nagata K et al: Plasma resistin concentration determined by common variants in the resistin gene and associated with metabolic traits in an aged Japanese population. Diabetologia 2009; 53: 234–246.
Yamada Y, Izawa H, Ichihara S et al: Prediction of the risk of myocardial infarction from polymorphisms in candidate genes. N Engl J Med 2002; 347: 1916–1923.
Heid IM, Wagner SA, Gohlke H et al: Genetic architecture of the APM1 gene and its influence on adiponectin plasma levels and parameters of the metabolic syndrome in 1727 healthy Caucasians. Diabetes 2006; 55: 375–384.
Menzaghi C, Ercolino T, Di Paola R et al: A haplotype at the adiponectin locus is associated with obesity and other features of the insulin resistance syndrome. Diabetes 2002; 51: 2306–2312.
Sutton BS, Weinert S, Langefeld CD et al: Genetic analysis of adiponectin and obesity in Hispanic families: the IRAS family study. Hum Genet 2005; 117: 107–118.
Hivert M-F, Manning AK, McAteer JB et al: Common variants in the adiponectin gene (ADIPOQ) associated with plasma adiponectin levels, type 2 diabetes, and diabetes-related quantitative traits: the Framingham Offspring study. Diabetes 2008; 57: 3353–3359.
Heid IM, Henneman P, Hicks A et al: Clear detection of ADIPOQ locus as the major gene for plasma adiponectin: results of genome-wide association analyses including 4659 European individuals. Atherosclerosis 2010; 208: 412–420.
Gu HF, Abulaiti A, Ostenson C-G et al: Single nucleotide polymorphisms in the proximal promoter region of the adiponectin (APM1) gene are associated with type 2 diabetes in Swedish Caucasians. Diabetes 2004; 53: S31–S35.
Mackevics V, Heid IM, Wagner SA et al: The adiponectin gene is associated with adiponectin levels but not with characteristics of the insulin resistance syndrome in healthy Caucasians. Eur J Hum Genet 2006; 14: 349–356.
Chow W-S, Cheung BMY, Tso AWK et al: Hypoadiponectinemia as a predictor for the development of hypertension: a 5-Year prospective study. Hypertension 2007; 49: 1455–1461.
Furuhashi M, Ura N, Higashiura K et al: Blockade of the renin-angiotensin system increases adiponectin concentrations in patients with essential hypertension. Hypertension 2003; 42: 76–81.
Ohashi K, Kihara S, Ouchi N et al: Adiponectin replenishment ameliorates obesity-related hypertension. Hypertension 2006; 47: 1108–1116.
Acknowledgements
We thank the study participants as well as Masakazu Kobayashi, Ken Harada, Takashi Yamada, Kazumasa Unno, Hiroki Kataoka, Hidehito Funahashi, Kousuke Arai, Yasutsugu Morimoto, Mayo Sukegawa, and especially Takao Nishizawa for their assistance. This study was supported in part by Grants-in-Aid for Scientific Research to MY including those of Categories (A) and (B) from the Japan Society for the Promotion of Science (17209021 and 21390209) and of Priority Area ‘Applied Genomics’ (1601223, 17019028, 18018020, and 20018026) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
ClinicalTrials.gov Identifiers
KING study: NCT00262691(http://clinicaltrials.gov/ct2/show/study/NCT00262691).
Replication study: NCT00408824 (http://clinicaltrials.gov/ct2/show/study/NCT00408824).
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Tanimura, D., Shibata, R., Izawa, H. et al. Relation of a common variant of the adiponectin gene to serum adiponectin concentration and metabolic traits in an aged Japanese population. Eur J Hum Genet 19, 262–269 (2011). https://doi.org/10.1038/ejhg.2010.201
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2010.201
Keywords
This article is cited by
-
ADIPOQ gene is linked to emotional eating behaviour in young Nigerian adults independent of psychological traits
Bulletin of the National Research Centre (2020)
-
Epigenome-wide association of myocardial infarction with DNA methylation sites at loci related to cardiovascular disease
Clinical Epigenetics (2017)
-
Polymorphisms of the adiponectin gene in gestational hypertension and pre-eclampsia
Journal of Human Hypertension (2014)
-
Combinational risk factors of metabolic syndrome identified by fuzzy neural network analysis of health-check data
BMC Medical Informatics and Decision Making (2012)