Abstract
RNA interference (RNAi) has been successfully employed for specific inhibition of gene expression; however, safety and delivery of RNAi remain critical issues. We investigated the combinatorial use of RNAi and U1 interference (U1i). U1i is a gene-silencing technique that acts on the pre-mRNA by preventing polyadenylation. RNAi and U1i have distinct mechanisms of action in different cellular compartments and their combined effect allows usage of minimal doses, thereby avoiding toxicity while retaining high target inhibition. As a proof of concept, we investigated knockdown of the firefly luciferase reporter gene by combinatorial use of RNAi and U1i, and evaluated their inhibitory potential both in vitro and in vivo. Co-transfection of RNAi and U1i constructs showed additive reduction of luciferase expression up to 95% in vitro. We attained similar knockdown when RNAi and U1i constructs were hydrodynamically transfected into murine liver, demonstrating for the first time successful in vivo application of U1i. Moreover, we demonstrated long-term gene silencing by AAV-mediated transduction of murine muscle with RNAi/U1i constructs targeting firefly luciferase. In conclusion, these results provide a proof of principle for the combinatorial use of RNAi and U1i to enhance target gene knockdown in vivo.
Similar content being viewed by others
Introduction
Gene silencing through RNA interference (RNAi) holds great potential for the treatment of various diseases, such as cancer, neurodegenerative disorders and viral infections. Since its basic discovery, RNAi research has advanced rapidly and has already made its way to the clinic, with phase I and II clinical trials under way.1, 2 RNAi is an evolutionarily conserved sequence-specific post-transcriptional gene silencing mechanism in eukaryotes that results in the degradation of mRNA and subsequently in decrease of protein synthesis.3, 4 Small double-stranded RNA molecules with perfect sequence complementarity to the target mRNA are processed by Dicer, rendering a mature small interfering RNA (siRNA) to interact with the cytosolic multiprotein RNA-induced silencing complex (RISC). The RISC-incorporated siRNA binds to a complementary mRNA sequence, which leads to cleavage of the target mRNA, followed by degradation through cellular RNases.5, 6
RNAi delivery methods employ both synthetic small interfering RNAs (siRNAs) that can be added to the cell, and DNA-based vectors that encode short hairpin RNAs (shRNAs), which upon expression are processed intracellularly to siRNAs.6 Expression of these shRNAs is typically regulated by constitutive strong polymerase III (pol III) promoters (H1 or U6), but their expression can also be made tissue specific or inducible, depending on the choice of promoter.7, 8 Specificity can be increased by encapsidation of the shRNA into a viral vector with specific tissue tropism. In addition, viral vector-mediated delivery of shRNA-encoding constructs has the advantage of long-term RNAi upon single administration.6, 9 Recombinant adeno-associated (AAV) viral vectors are excellent vehicles for RNAi-based gene therapy as they allow efficient, safe, long-term gene delivery in a wide range of tissues. AAV is a single-stranded DNA virus with a 4.7-kb genome flanked by inverted terminal repeats (ITRs), which are cis-acting elements needed for AAV packaging and DNA replication. Recombinant AAV is devoid of all viral genes except the ITR sequences and is considered a safe vector as it lacks pathogenicity and persists mainly episomally, rather than integrating in the host genome. AAV vectors have great versatility and delivery potential through the availability of multiple serotypes, which can be expanded further by pseudotyping and capsid modification.10, 11
Despite successful demonstration of RNAi in vivo and initiation of clinical trials using RNAi-based therapeutics, safety and delivery of RNAi remain critical issues.1, 12 Adverse immune responses, off-target effects and saturation of the cellular RNAi processing machinery have been reported, which demonstrates the need for optimization of therapeutic gene-silencing technologies.13, 14, 15 For gene therapy-based RNAi, this could be achieved by controlling intracellular shRNA expression levels, which however requires cumbersome optimization. An alternative approach is to combine RNAi with gene-silencing technologies with a different mode of action that do not use the same processing pathway as RNAi.
U1 interference (U1i) is a novel post-transcriptional gene-silencing method that inhibits polyadenylation and maturation of pre-mRNAs. The technology is based on a modified U1 small nuclear ribonucleoprotein particle (snRNP). Mammalian wild-type U1 snRNP consists of a 164-nt U1 small nuclear RNA (snRNA) bound by 10 polypeptides, one among which is U1-70K, and it functions as part of the spliceosome in pre-mRNA splicing by hybridizing to the 5′ splice sites of introns using the 10 nucleotides at its 5′ end for recognition. In addition, U1 snRNP has an alternative role in the inhibition of gene expression by binding the 3′ untranslated region of papillomaviruses and certain mammalian genes.16, 17, 18 Upon base-pairing of the 10 nucleotides at the 5′ end of U1 snRNA to a target sequence in the 3′ untranslated region, polyadenylation of the target pre-mRNA is inhibited through a direct interaction of the U1-70K protein subunit of U1 snRNP with poly(A) polymerase.19 Inhibition of polyadenylation prevents maturation of pre-mRNA, which is subsequently degraded in the nucleus. By using this naturally occurring mechanism, a gene-specific silencing method based on inhibition of pre-mRNA polyadenylation and maturation is established. Specific modification of the 10 nucleotides at the 5′ end of U1 snRNA leads to base-pairing with the 3′ terminal exon of a desired target gene, resulting in the inhibition of polyadenylation, prevention of maturation, and finally in targeting of pre-mRNA for degradation. This approach has been successfully employed in vitro upon transient and stable delivery of DNA-encoding modified U1 snRNA to various cell lines, resulting in the repression of reporter genes as well as endogenous genes.20, 21, 22, 23, 24, 25, 26
Combining RNAi and U1i gene silencing yields more efficient knockdown of gene expression, as these methods have distinct mechanisms of action in different cellular compartments.27 RNAi takes place in the cytosol and involves cleavage of mRNA, whereas U1i is effective in the nucleus and functions through prevention of pre-mRNA maturation.6, 19 A combinatorial approach allows usage of minimal doses of individual components (RNAi and U1i), thereby reducing the chances of toxicity, such as saturation of cellular RNAi pathways. Simultaneous application of siRNA and U1i molecules that target the same gene has been shown to enhance silencing in vitro.18 Furthermore, Abad et al.27 recently demonstrated synergistic enhancement of reporter and endogenous gene silencing by the combined action of RNAi with U1i. However, the effectiveness of these combined silencing methods has yet to be demonstrated in vivo.
Here, we employed the firefly luciferase reporter gene to quantify gene silencing by the combined action of RNAi and U1i. First, shRNA and U1i plasmids targeting firefly luciferase were co-transfected in vitro in HEK293T cells. Second, the in vivo activity of co-transfected shRNA and U1i plasmids was assessed in murine liver using hydrodynamic transfection. Finally, AAV-mediated transduction of murine muscle was employed to demonstrate stable in vivo suppression of luciferase expression by combinatorial RNAi and U1i. We show that co-transfection of RNAi with U1i constructs has an additive effect on reporter gene knockdown both in vitro and in vivo, compared with transfection with either effector alone. In addition, AAV-delivered RNAi and U1i result in prolonged inhibition of luciferase expression in murine muscle. Overall, these data illustrate the effectiveness of RNAi and U1i gene-silencing methods both in vitro and in vivo, which is of interest for the development of therapeutic applications.
Results
Screening of shRNA and U1i constructs targeting luciferase
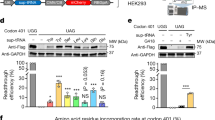
To evaluate the inhibitory effect of RNAi and U1i on gene expression, we chose the firefly luciferase reporter as a target gene. Knockdown of luciferase was assayed by transfecting shRNA constructs targeting firefly luciferase together with firefly and renilla luciferase reporter plasmids. The ratio of firefly and renilla luciferase signal was used to calculate relative luciferase activity. shLuc1 was highly effective and showed a dose-dependent luciferase knockdown up to approximately 85%, whereas shLuc2 was ineffective (Figure 1a).
In vitro knockdown of luciferase by shRNA and U1i constructs. (a) Increasing amounts (1–250 ng) of shRNA constructs targeting firefly luciferase (shLuc1 and shLuc2) were co-transfected with 2.5 ng firefly luciferase and 0.5 ng renilla luciferase plasmid in 96-well plates containing approximately 2.5 × 104 HEK293T cells. The total amount of transfected DNA was kept constant by adding the pSuper cloning vector. At 2 days after transfection, cells were lysed and firefly and renilla luciferase signals were measured. Relative luciferase activity was calculated as the ratio of firefly and renilla luciferase and plotted relative to the 100% control transfected with 250 ng pSuper. shLuc1 significantly reduced luciferase by 85%, whereas shLuc2 was inactive. Control shRNA plasmids were shGFP and shApoB. (b) Co-transfection of 200 ng U1i constructs targeting firefly luciferase (L1-L8) with firefly luciferase and renilla luciferase plasmid. Cells transfected with 200 ng control plasmid U1C were set at 100%. L4 and L5 mildly reduced luciferase expression by 30 and 40%, respectively, compared with U1C. (c) Co-transfection of 100 ng L4 and 100 ng L5 significantly improved luciferase inhibition, compared with transfection with 200 ng L4 or L5 alone (P<0.05). Data are presented as mean of three technical replicates±s.d.
Eight U1 interference (U1i) constructs targeting firefly luciferase (L1–L8) were designed and screened for knockdown of firefly luciferase. Co-transfection of L1–L8 constructs with firefly and renilla luciferase reporters demonstrated a mild inhibitory effect of two out of the eight U1i constructs (Figure 1b). L4 and L5 reduced luciferase expression by 30 and 40%, respectively. We proceeded to test the combined effect of the two mildly effective U1i constructs with other U1i constructs in order to screen for improved efficacy (Supplementary Table 1). The strongest decrease in luciferase expression was observed by the combination of L4 with L5 and reached around 80% (Supplementary Table 1 and Figure 1c). Next, a bicistronic expression vector was cloned that simultaneously expressed L4 and L5 from one backbone. Transfection with this double U1i plasmid L4/L5 was equally effective in suppressing luciferase activity as co-transfection with L4 and L5 single plasmids, indicating that there is efficient expression of both effectors from one backbone (data not shown). Thus, although the single U1i constructs tested were only mildly effective on their own, they can be combined to reduce luciferase expression to a similar level as the shLuc1 construct.
Co-expression of shRNA and U1i enhances luciferase knockdown in vitro
Combining RNAi and U1i gene silencing allows more efficient knockdown of gene expression as they have distinct mechanisms of action in different cellular compartments. Therefore, we co-transfected shLuc1 together with the most effective U1i constructs L4 and L5. Transfection of L4 or L5 combined with control shApoB led to a mild decrease in luciferase expression of approximately 30 and 40%, respectively, comparable to the single U1i constructs (Figures 1b and 2a). Expression of the control plasmid shApoB together with the combined expression of L4 and L5 or the bicistronic L4/L5 construct suppressed luciferase even more (Figure 2a). When shLuc1 was co-transfected with control U1C, an 86% reduction in luciferase expression was observed, similar to the inhibition shown with shLuc1 alone (Figures 1a, 2a and b). We were able to significantly enhance luciferase knockdown when the three luciferase-targeting constructs, shLuc1, L4, and L5, were combined and when shLuc1 was combined with the bicistronic L4/L5 construct, resulting in a 95% reduction in luciferase activity (Figure 2b). The additive effect of L4 and L5 was shown for several shLuc1 concentrations (Supplementary Figure 1). Hence, co-expression of RNAi and U1i enhances target gene knockdown in vitro, consistent with previous data.27
Combination of RNAi and U1i increases luciferase knockdown in vitro. (a) Co-transfection of 50 ng control shApoB construct or 50 ng shLuc1 construct with 200 ng (or 100+100 ng) U1i plasmid and firefly luciferase and renilla luciferase plasmid. Transfections and analysis were carried out as described in Figure 1a. Cells transfected with a combination of 50 ng shApoB and 200 ng U1C were set at 100%. Co-transfection of shApoB with L4 and L5 or L4/L5 significantly reduced luciferase activity by ∼65% compared with the 100% control (P<0.05). Transfection of shLuc with U1C resulted in a significant 86% knockdown (P<0.001). (b) Co-transfection of 50 ng shLuc1 construct with 200 ng (or 100+100 ng) U1i plasmid and firefly luciferase and renilla luciferase. Note the different scales on the y axes in (a, b). Co-transfection of shLuc1 with L4 and L5 maximally reduced luciferase expression by 95%. Control U1i constructs were U1C, and H1 and H4 that target the human huntingtin sequence. Luciferase activity after co-transfection of shLuc1 with L4 and L5 or L4/L5 was significantly different from co-transfection of shLuc1 with U1C (P<0.05), and from co-transfection of shLuc1 with H1 and H4 (P<0.001) or H1/H4 (P<0.05). Data are presented as mean of three independent experiments +s.d., analyzed using factor correction.36
Transient in vivo delivery of shRNA and U1i enhances luciferase knockdown in murine liver
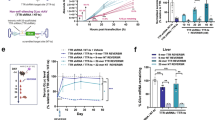
The in vivo silencing activity of RNAi and U1i was evaluated in murine liver by hydrodynamic co-transfection of shLuc1 and bicistronic L4/L5 plasmids. shLuc1 was mixed with L4/L5 and injected together with the firefly luciferase reporter plasmid and a construct expressing secreted alkaline phosphatase (SEAP). Plasma measurements of co-injected SEAP plasmid served to normalize for transfection efficiency. In vivo delivery of shLuc1 and L4/L5 plasmids resulted in 85% knockdown of relative luciferase expression at two days post-transfection, which was comparable to luciferase knockdown observed in vitro (Figures 2b and 3). In addition, we observed intermediate suppressive effects of L4/L5 alone (25% in combination with control shApoB) and of shLuc1 alone (61% in combination with control H1/H4) (Figure 3). These results demonstrate the feasibility of in vivo suppression of a target gene by the combined action of RNAi and U1i.
Hydrodynamic delivery of shRNA and U1i constructs to murine liver inhibits luciferase activity in vivo. Mice were hydrodynamically co-transfected with 10 μg shRNA (shLuc1 or control shApoB) and 40 μg double U1i (L4/L5 or control H1/H4) plasmid and with 2 μg firefly luciferase reporter and 0.5 μg secreted alkaline phophatase (SEAP) plasmid. Both the firefly luciferase and SEAP reporter gene were expressed under control of the liver-specific AAT promoter, as hydrodynamically delivered plasmid DNA localizes mainly to the liver. Two days after transfection, bioluminescence was measured in the IVIS, and luciferase signal was calculated relative to plasma SEAP levels. Murine liver co-transfected with control constructs shApoB and H1/H4 was set at 100%. Co-transfection of shLuc1 with L4/L5 maximally reduced luciferase activity by 84%. However, luciferase knockdown was not significantly different between treatments. Data are presented as mean (n=4–8)±s.e.
Simultaneous expression of shRNA and bicistronic U1i constructs from a single backbone inhibits luciferase expression in vitro
In order to develop therapeutic applications using RNAi and U1i, durable expression of these gene-silencing mechanisms has to be established. For long-term expression, AAV vectors are excellent delivery vehicles that are capable of transducing a wide range of tissues with high efficiency. Therefore, we proceeded to express shLuc1 and L4/L5 from an AAV backbone. The U1i bicistronic construct L4/L5 was cloned together with shLuc1 in a pro-AAV cloning vector (LL) (Supplementary Figure 2). Each individual effector was expressed from their own promoter and termination sequences. Three additional vectors were made: one that expressed control shApoB and control U1i H1/H4 (AH), a second that expressed anti-luciferase U1i L4/L5 and control shApoB (AL), and a third that expressed anti-luciferase shLuc1 and control U1i H1/H4 (LH). Upon transfection of HEK293T cells, qPCR analysis confirmed that all four shRNA/U1i constructs expressed recombinant U1 snRNA (Supplementary Figure 3a). Furthermore, their expression did not affect cell viability, as determined by the MTT toxicity assay (Supplementary Figure 3b). To assess the in vitro inhibitory potential of the shRNA/U1i co-expression constructs, increasing amounts of plasmid were co-transfected together with firefly and renilla reporters. All luciferase-targeting constructs (LH, AL, LL) significantly suppressed luciferase activity compared with the control AH (Figure 4). Of these, the RNAi and U1i double inhibitory construct (LL) was most effective and reduced luciferase expression by 85%. Although the combined effect of shLuc1 and L4/L5 (LL) appeared to be stronger than the effect of shLuc1 alone (LH), this effect was not significant. This may be due to the fact that all three constructs were expressed from one vector, thereby missing the ability to differentially dose the inhibitory constructs for optimal effect.
Co-expression of RNAi and U1i from one vector backbone suppresses luciferase activity in vitro. Increasing amounts (25–250 ng) of shRNA/U1i plasmid expressing both shRNA and double U1i constructs were co-transfected with firefly luciferase and renilla luciferase plasmid and analyzed as described in Figure 1a. The total amount of transfected DNA was kept constant by adding the pro-AAV cloning vector. Cells transfected with 250 ng pro-AAV vector were set at 100%. The control vector AH expressed shApoB and double H1/H4. The U1i vector AL expressed shApoB and double L4/L5. The shRNA vector LH expressed shLuc1 and double H1/H4. The shRNA and U1i combination vector LL expressed shLuc1 and double L4/L5. AL, LH, and LL significantly reduced luciferase activity compared with the AH control vector (P<0.001) and LL was most effective and led to 85% luciferase knockdown. Data are presented as mean of three independent experiments±s.d., analyzed using factor correction.36
AAV-mediated knockdown of luciferase using RNAi and U1i in murine muscle
Having established the in vitro efficacy of the shRNA/U1i co-expression constructs, we proceeded to investigate long-term in vivo suppression of luciferase in murine muscle by employing AAV serotype 1 viral vectors to deliver shLuc1 and L4/L5. AAV1-expressing luciferase reporter (AAV-Luc) was co-injected intramuscularly with the AAV viral vectors encoding the shRNA/U1i co-expression constructs AAV-AH (shApoB and H1/H4), AAV-AL (shApoB and L4/L5), AAV-LH (shLuc1 and H1/H4), or AAV-LL (shLuc1 and L4/L5). All AAV-shRNA/U1i vectors co-expressed the GFP reporter gene, and full muscle transduction was demonstrated by monitoring GFP expression at 8 weeks post-injection (data not shown). Recombinant U1 snRNA expression was shown for all four AAV-shRNA/U1i vectors in transduced muscle at 8 weeks post-transduction (Supplementary Figure 4). In addition, the absence of muscle damage was demonstrated by creatine phosphokinase (CPK) assay and by hematoxylin and eosin (H&E) staining of frozen muscle sections (data not shown).
Luciferase expression was monitored at 1, 2, 3, 4, 6, and 8 weeks post-transduction (Figure 5). Overall, luciferase expression increased over time during the first weeks post-transduction with AAV-Luc and AAV-shRNA/U1i, and reached a relatively stable level at 4 weeks, characteristic of AAV expression kinetics.28 Transduction with AAV-shRNA/U1i targeting firefly luciferase (AAV-LL) reduced luciferase expression significantly by 25–40% compared with the control vector AAV-AH (Figure 5). This effect was already observed at 1 week post-transduction and remained stable throughout the course of the experiment, demonstrating the effectiveness and stability of luciferase knockdown by combined RNAi and U1i expression in vivo. Overall, luciferase suppression in vivo was lower than observed in vitro; the maximum inhibitory effect of shLuc and L4/L5 (LL) was 40% in vivo, versus 85% in vitro (Figures 4 and 5). Moreover, we could not observe a significant effect of the U1i constructs alone (AAV-AL), or of the shRNA construct alone (AAV-LH). The fact that we did not observe an intermediate inhibitory effect of these constructs may reflect an overall lower knockdown in vivo compared with the effect measured in vitro (Figures 4 and 5). In conclusion, we demonstrated long-term stable transduction of murine muscle using AAV delivery vectors and observed sustained in vivo knockdown of a luciferase reporter by the combined action of RNAi and U1i.
AAV-mediated in vivo knockdown of luciferase using RNAi and U1i. Murine muscle was co-transduced with 1 × 1010 gc AAV-Luc and 4.5 × 1010 gc AAV-shRNA/U1i per hind limb. The gastrocnemic and adductor muscles of both hind limbs were injected. Six mice per treatment were injected, resulting in n=12. Luciferase expression was measured in the IVIS at 1, 2, 3, 4, 6, and 8 weeks post-transduction. The control vector AAV-AH expressed shApoB and double H1/H4. The U1i vector AAV-AL expressed shApoB and double L4/L5. The shRNA vector AAV-LH expressed shLuc1 and double H1/H4. The shRNA and U1i combination vector AAV-LL expressed shLuc1 and double L4/L5, and this viral vector significantly reduced luciferase expression on average by 25% during the course of the experiment (P<0.05). Data are presented as mean (n=12)±s.e.
Discussion
Delivery and safety of the therapeutic molecules remain major challenges in the RNAi field. These safety concerns involve unwanted activation of the immune response, off-targeting effects, and saturation of the cellular RNAi machinery due to overexpression of the shRNA.13, 14, 15 To minimize the risk of these unwanted effects while retaining high target inhibition, we investigated the combined action of RNAi and U1i, two gene-silencing mechanisms with different modes of action. As a proof of concept, we studied the inhibitory effect of RNAi and U1i on luciferase reporter expression, and demonstrated increased luciferase knockdown in vitro by shRNA and U1i transfection as well as prolonged suppression in vivo using AAV-delivered shRNA/U1i expression cassettes.
Combinatorial RNAi strategies that aim to avert viral resistance (for example, of HIV) by triggering RNAi against multiple targets have been previously developed. These studies often involve expression of several shRNA molecules, which is expected to proportionally increase the toxicity and off-targeting risk associated with mono-RNAi treatments.29 To circumvent these problems, Hemmings-Mieszczak et al.30 combined siRNAs with antisense oligonucleotides and observed synergistic knockdown effects in cultured cells. Similarly, Jarczak et al.31 mixed shRNAs with hammerhead ribozymes, which overall increased target gene inhibition. The in vivo efficacy of these combinatorial approaches nevertheless remains to be demonstrated. Combinatorial use of RNAi with U1i in cell lines has been described previously for both reporter and endogenous genes, and was shown to yield synergistic suppressive effects on target gene expression.18, 27 We observed a similar effect when transfecting shLuc1 with two U1i constructs L4 and L5, which resulted in 95% inhibition of luciferase activity. Of the individual components, shLuc1 was most effective, yielding ∼85% knockdown, and the combination of L4 with L5 resulted in ∼70–80% luciferase inhibition. Previously, increased target knockdown was observed when several U1i constructs were expressed that recognized multiple targets, or when a U1i construct was expressed that recognized repeat regions in the 3′ terminal exon of a target gene.22 However, the U1i constructs tested here were only mildly effective on their own. When screening the eight U1i constructs (L1–L8), we observed not only inhibition of luciferase expression, but even stimulatory effects on luciferase expression by two of the U1i constructs, L3 and L6; the cause for this is currently unclear. Successful design of U1i constructs seems to depend most strongly on the accessibility of the secondary structure of the target region and may have been suboptimal.26 In addition, Abad et al.27 observed a milder inhibitory effect of their U1i construct when the target sequence was expressed within the firefly luciferase plasmid, compared with expression in the previously used renilla luciferase construct, and suggested that this decrease in efficacy might reflect the effect of neighboring sequences.
To prove the feasibility of combinatorial RNAi and U1i, we aimed at demonstrating luciferase knockdown in vivo upon delivery of shRNA and U1i constructs in a murine model. U1i has not been established in animal models yet, and this is considered an important step in the development of this gene-silencing mechanism as a therapy.32 Two approaches were employed, each targeting a different organ; transient luciferase knockdown by plasmid delivery of shLuc1 and L4/L5 to the liver, and their long-term effect by AAV-mediated expression in the muscle. A strong reduction in luciferase expression was shown upon hydrodynamic transfection of murine liver with shLuc1 and the bicistronic L4/L5 construct, and intermediate inhibitory effects were observed after transfection with shLuc1 and L4/L5 alone. These results matched the observations in cell cultures, and prompted us to investigate the long-term silencing ability of combinatorial RNAi and U1i in murine muscle. AAV-mediated transduction of murine muscle is expected to yield high-level long-term expression, and indeed, the AAV-delivered vector co-expressing shLuc1 and double L4/L5 significantly reduced luciferase expression compared with the control AAV vector. However, the effect was relatively mild compared with in vitro observations. In addition, no significant luciferase suppression by shLuc1 or L4/L5 alone was measured. Possibly, the lower efficacy is due to the fact that all three effectors were expressed from one backbone, which might result in promoter interference when expressed in vivo. Alternatively, co-injection of AAV-Luc with AAV-shRNA/U1i viral vectors may have resulted in transduced cells that express luciferase, but not the shRNA/U1i inhibitors, or vice versa. This would lower the overall efficacy of luciferase knockdown. Nevertheless, the inhibitory effect of AAV-delivered shLuc and L4/L5 lasted throughout the course of the 8-week experiment and failed to show any obvious signs of toxicity, as determined by CPK assay and H&E staining. Thus, although the AAV vectors and possibly the AAV dose need to be optimized, we were able to show a proof of principle for the in vivo use of combinatorial RNAi and U1i.
Future research is needed to optimize the inhibitory potential of combined RNAi and U1i in vivo when delivered with AAV, and furthermore requires investigation into the potential toxic or off-target effects of U1i gene silencing. With respect to the latter, U1i has the advantage that it only works in the 3′ terminal exon of a target gene, and that secondary structures occlude target sequences.22, 26 Indeed, Goraczniak et al.18 reported comparable limited off-target effects by siRNA and U1 snRNA-mediated gene silencing in microarray analyses. In addition, the naturally occurring U1 snRNP is highly abundant, with about one million copies present in a typical mammalian nucleus, and therefore overexpression of modified U1i constructs is not expected to interfere with the overall splicing machinery.33 Furthermore, stable expression of U1i constructs has been demonstrated in stably transduced cell lines, thus indicating the low toxicity of this technique.22 Finally, optimization and validation of U1i design rules should improve target gene knockdown, which allows use of lower dose of U1i constructs, thus aiding in improving the safety of the U1i gene-silencing technique. In conclusion, this study shows for the first time the in vivo application of U1i and furthermore demonstrates the in vivo efficacy of combinatorial RNAi and U1i.
Materials and methods
DNA constructs
shRNA constructs were made by annealing complementary oligonucleotides and ligating them into the BglII and XhoI site of the pSuper vector (OligoEngine, Seattle, WA, USA). shRNAs expressed from this plasmid are transcribed from a H1 promoter. Two shRNAs were designed that target firefly luciferase (shLuc1 and shLuc2) and two control shRNAs were made that target GFP or human ApoB (shGFP and shApoB). The sequences of the oligonucleotides used in this study are listed in Supplementary Table 2. Eight U1i constructs were designed that target firefly luciferase (L1–L8), and three control U1i constructs were made that target human huntingtin (H1 and H4) or express the wild-type U1 sequence (U1C).22 The locations of each shRNA and U1i luciferase target sequence are shown in Supplementary Figure 5. U1i constructs were made by annealing complementary oligonucleotides and ligating them into the BglII and BclI sites of a pGEM-3Zf+ plasmid (Promega, Madison, WI, USA) encoding the U1 snRNA sequence including promoter and termination sequences.26 Bicistronic U1i constructs encoding L4/L5 and H1/H4 were made by digestion of L4 and H4 with XbaI, followed by blunting of these digested plasmids. Next, the constructs were digested with EcoRI. The resulting double-digested L4 and H4 were then ligated into the blunt SmaI site and EcoRI site of L5 and H1, respectively. Selected shRNA (shLuc1 and shApoB) and U1i double (L4/L5 and H1/H4) constructs were further subcloned into a pro-AAV vector that encodes CMV-eGFP, using SphI (for shRNA), and MunI and SalI (for double U1i constructs) (Supplementary Figure 2). The resulting AAV-shRNA/U1i plasmids are AH (shApoB/H1/H4), AL (shApoB/L4/L5), LH (shLuc1/H1/H4), and LL (shLuc1/L4/L5). Reporter constructs that were used are pRL-CMV-renilla (Promega), and firefly luciferase pGL4 (Promega) under control of the CMV promoter or the liver-specific AAT promoter in a pGEM-3Zf+ plasmid (Promega). Plasmid encoding AAT-secreted alkaline phosphatase (AAT-SEAP) was made by BaseClear (Leiden, The Netherlands).
AAV vector production
AAV serotype 1 vectors were produced by calcium phosphate-mediated co-transfection in human embryonic kidney (HEK) 293 cells as described previously.34 Viral batches were produced for AAV-shRNA/U1i constructs AH, AL, LH, and LL and crude lysate was purified by affinity chromatography, diafiltrated in 5% PBS sucrose (v/w) by cross-flow filtration and further concentrated using centricon tubes (YM-100; Millipore, Bedford, MA, USA). The final concentration of 5–10 × 1011 genome copies per ml was determined by qPCR (Applied Biosystems, Foster City, CA, USA) using primers pr59 and pr60 (Supplementary Table 2), amplifying a 70-bp fragment from the CMV promoter region. AAV-CMV-luciferase (AAV-Luc) was produced in insect cells through the use of recombinant baculovirus technology, as described in Smith et al.35
Luciferase assay
The HEK293T cell line was maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA, USA) containing 10% fetal calf serum, 100 U ml−1 penicillin, and 100 U ml−1 streptomycin at 37 °C and 5% CO2. For luciferase assays, approximately 2.5 × 104 HEK293T cells were seeded in 96-well plates and transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Cells were co-transfected with 2.5 ng CMV-firefly luciferase plasmid and 0.5 ng pRL-CMV-renilla luciferase plasmid, together with a selection of shRNA (1–250 ng), U1i (100–200 ng), and shRNA/U1i (25–250 ng) constructs. The total amount of transfected DNA was kept constant per experiment by adding vector control plasmid (pSuper, pGEM-3Zf+, or pro-AAV plasmid). Two days after transfection, firefly and renilla luciferase expression was assessed using a luminometer (Spectramax L; Molecular Devices, Sunnyvale, CA, USA) according to the Dual-Luciferase Reporter Assay System (Promega). Relative luciferase activity was calculated as the ratio between firefly and renilla luciferase activities. To correct for between-session variation in replicate experiments, the factor correction program was used.36 Statistical analysis was performed using one-way analysis of variance, with or without repeated measures, and a Bonferroni post hoc comparison. A P-value <0.05 was considered significant.
In vivo transfection of murine liver using hydrodynamic tail vein injection
All animal experiments were conducted according to the guidelines of the local animal welfare committee. Six-to-eight-week-old female C57BL/6 mice were anesthetized using intraperitoneal injection with fentanyl-fluanisone-midazolam (FFM) mix and hydrodynamically transfected via the tail vein with plasmids encoding AAT-Luc (2 μg), AAT-SEAP (0.5 μg), shRNA (shApoB or shLuc1) (10 μg), and double U1i (H1/H4 or L4/L5) (40 μg) in 1.5 ml Ringer's solution (0.9% NaCl, 0.03% KCl, and 0.016% CaCl2). Two days after transfection, luciferase was measured in living mice in the Spectrum In Vivo Imaging System (IVIS, Caliper Life Sciences, Hopkinton, MA, USA). Mice were anesthetized with 2% isoflurane and injected intraperitoneally with 3 mg D-luciferin (Synchem, Felsberg, Germany) in 200 μl PBS. Regions of interest (ROIs) were traced over the positions of greatest signal intensity on the animal and light intensity was quantified as photons s−1 cm−2 sr−1. At the same time point, plasma was collected by retro-orbital bleed and secreted alkaline phosphatase (SEAP) was measured using the chemiluminescent SEAP reporter gene assay according to the manufacturer's instructions (Roche Diagnostics GmbH, Mannheim, Germany). Luciferase readings were normalized to SEAP levels to adjust for transfection efficiency. Statistical analysis was performed using one-way analysis of variance and a Bonferroni post hoc comparison. A P-value <0.05 was considered significant.
In vivo transduction of murine muscle using AAV
Six-to-eight-week-old female Balb/C mice were injected intramuscularly with a mixture of AAV-Luc and AAV-shRNA/U1i (AAV-AH, AAV-AL, AAV-LH, or AAV-LL). Gastrocnemic and adductor muscles were injected with 1 × 1010 gc per limb AAV-Luc and 4.5 × 1010 gc per limb AAV-shRNA/U1i. Luciferase expression in living mice was monitored in the IVIS at 1, 2, 3, 4, 6, and 8 weeks post-transduction, as described above. Statistical analysis was performed using one-way analysis of variance with repeated measures and a Bonferroni post hoc comparison. A P-value <0.05 was considered significant.
References
Castanotto D, Rossi JJ . The promises and pitfalls of RNA-interference-based therapeutics. Nature 2009; 457: 426–433.
Tiemann K, Rossi JJ . RNAi-based therapeutics—current status, challenges and prospects. EMBO Mol Med 2009; 1: 142–151.
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC . Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998; 391: 806–811.
Meister G, Tuschl T . Mechanisms of gene silencing by double-stranded RNA. Nature 2004; 431: 343–349.
Zamore PD, Tuschl T, Sharp PA, Bartel DP . RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21–23 nucleotide intervals. Cell 2000; 101: 25–33.
Grimm D . Small silencing RNAs: state-of-the-art. Adv Drug Deliv Rev 2009; 61: 672–703.
Giering JC, Grimm D, Storm TA, Kay MA . Expression of shRNA from a tissue-specific pol II promoter is an effective and safe RNAi therapeutic. Mol Ther 2008; 16: 1630–1636.
Laurenti E, Barde I, Verp S, Offner S, Wilson A, Quenneville S et al. Inducible gene and shRNA expression in resident hematopoietic stem cells in vivo. Stem Cells 2010; 28: 1390–1398.
Grimm D, Pandey K, Kay MA . Adeno-associated virus vectors for short hairpin RNA expression. Methods Enzymol 2005; 392: 381–405.
Le Bec C, Douar AM . Gene therapy progress and prospects—vectorology: design and production of expression cassettes in AAV vectors. Gene Therapy 2006; 13: 805–813.
Daya S, Berns KI . Gene therapy using adeno-associated virus vectors. Clin Microbiol Rev 2008; 21: 583–593.
Grimm D, Kay MA . RNAi and gene therapy: a mutual attraction. Hematology 2007; 2007: 473–481.
Judge A, MacLachlan I . Overcoming the innate immune response to small interfering RNA. Hum Gene Ther 2008; 19: 111–124.
Rao DD, Senzer N, Cleary MA, Nemunaitis J . Comparative assessment of siRNA and shRNA off target effects: what is slowing clinical development. Cancer Gene Ther 2009; 16: 807–809.
Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 2006; 441: 537–541.
Furth PA, Choe WT, Rex JH, Byrne JC, Baker CC . Sequences homologous to 5′ splice sites are required for the inhibitory activity of papillomavirus late 3′ untranslated regions. Mol Cell Biol 1994; 14: 5278–5289.
Guan F, Caratozzolo RM, Goraczniak R, Ho ES, Gunderson SI . A bipartite U1 site represses U1A expression by synergizing with PIE to inhibit nuclear polyadenylation. RNA 2007; 13: 2129–2140.
Goraczniak R, Behlke MA, Gunderson SI . Gene silencing by synthetic U1 adaptors. Nat Biotech 2009; 27: 257–263.
Gunderson SI, Polycarpou-Schwarz M, Mattaj IW . U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol Cell 1998; 1: 255–264.
Beckley SA, Liu P, Stover ML, Gunderson SI, Lichtler AC, Rowe DW . Reduction of target gene expression by modified U1 snRNA. Mol Cell Biol 2001; 21: 2815–2825.
Liu P, Gucwa A, Stover ML, Buck E, Lichtler A, Rowe D . Analysis of inhibitory action of modified U1 snRNAs on target gene expression: discrimination of two RNA targets differing by a 1 bp mismatch. Nucl Acids Res 2002; 30: 2329–2339.
Fortes P, Cuevas Y, Guan F, Liu P, Pentlicky S, Jung SP et al. Inhibiting expression of specific genes in mammalian cells with 5′ end-mutated U1 small nuclear RNAs targeted to terminal exons of pre-mRNA. Proc Natl Acad Sci USA 2003; 100: 8264–8269.
Liu P, Kronenberg M, Jiang X, Rowe D . Modified U1 snRNA suppresses expression of a targeted endogenous RNA by inhibiting polyadenylation of the transcript. Nucl Acids Res 2004; 32: 1512–1517.
Sajic R, Lee K, Asai K, Sakac D, Branch DR, Upton C et al. Use of modified U1 snRNAs to inhibit HIV-1 replication. Nucl Acids Res 2007; 35: 247–255.
Jankowska A, Gunderson S, Andrusiewicz M, Burczynska B, Szczerba A, Jarmolowski A et al. Reduction of human chorionic gonadotropin beta subunit expression by modified U1 snRNA caused apoptosis in cervical cancer cells. Mol Cancer 2008; 7: 26.
Abad X, Vera M, Jung SP, Oswald E, Romero I, Amin V et al. Requirements for gene silencing mediated by U1 snRNA binding to a target sequence. Nucl Acids Res 2008; 36: 2338–2352.
Abad X, Razquin N, Abad A, Fortes P . Combination of RNA interference and U1 inhibition leads to increased inhibition of gene expression. Nucl Acids Res 2010; 38: e136.
Riviere C, Danos O, Douar AM . Long-term expression and repeated administration of AAV type 1, 2 and 5 vectors in skeletal muscle of immunocompetent adult mice. Gene Therapy 2006; 13: 1300–1308.
Grimm D, Kay MA . Combinatorial RNAi: a winning strategy for the race against evolving targets? Mol Ther 2007; 15: 878–888.
Hemmings-Mieszczak M, Dorn G, Natt FJ, Hall J, Wishart WL . Independent combinatorial effect of antisense oligonucleotides and RNAi-mediated specific inhibition of the recombinant Rat P2X3 receptor. Nucl Acids Res 2003; 31: 2117–2126.
Jarczak D, Korf M, Beger C, Manns MP, Kruger M . Hairpin ribozymes in combination with siRNAs against highly conserved hepatitis C virus sequence inhibit RNA replication and protein translation from hepatitis C virus subgenomic replicons. FEBS J 2005; 272: 5910–5922.
Grunweller A, Hartmann RK . Expanding RNA silencing approaches by U1 adaptors. ChemBioChem 2009; 10: 1599–1601.
Will CL, Luhrmann R . Protein functions in pre-mRNA splicing. Curr Opin Cell Biol 1997; 9: 320–328.
Blits B, Derks S, Twisk J, Ehlert E, Prins J, Verhaagen J . Adeno-associated viral vector (AAV)-mediated gene transfer in the red nucleus of the adult rat brain: comparative analysis of the transduction properties of seven AAV serotypes and lentiviral vectors. J Neurosci Methods 2010; 185: 257–263.
Smith RH, Levy JR, Kotin RM . A simplified baculovirus-AAV expression vector system coupled with one-step affinity purification yields high-titer rAAV stocks from insect cells. Mol Ther 2009; 17: 1888–1896.
Ruijter J, Thygesen H, Schoneveld O, Das A, Berkhout B, Lamers W . Factor correction as a tool to eliminate between-session variation in replicate experiments: application to molecular biology and retrovirology. Retrovirology 2006; 3: 2.
Acknowledgements
We thank Amaya Abad for technical assistance and Mark Chadwick and Wim Hermens for critically reviewing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Gene Therapy website
Supplementary information
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Koornneef, A., van Logtenstein, R., Timmermans, E. et al. AAV-mediated in vivo knockdown of luciferase using combinatorial RNAi and U1i. Gene Ther 18, 929–935 (2011). https://doi.org/10.1038/gt.2011.41
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gt.2011.41
Keywords
This article is cited by
-
In vivo knockdown of intersectin-1s alters endothelial cell phenotype and causes microvascular remodeling in the mouse lungs
Apoptosis (2013)
-
Optimization and comparison of knockdown efficacy between polymerase II expressed shRNA and artificial miRNA targeting luciferase and Apolipoprotein B100
BMC Biotechnology (2012)