Abstract
Adult height is a highly heritable, classic polygenic trait. The recent advent of genome-wide association studies has led to the identification of robust association at common variants influencing the normal variation of height; the number of loci has now risen to 50. The present study tested the potential overlap of height associations at 46 loci that were previously reported in European-descent populations with those among 1530 Japanese subjects and also attempted to replicate the suggestive association signals that were previously reported in East Asians alone. We found a total of 20 independent loci to be significantly (P<0.05) associated with height among the Japanese subjects (N⩽6814); 19 loci were originally identified in Europeans and the remaining 1 locus (ZFAT) was previously reported in Koreans (P=3.8 × 10−9 with the Japanese and Korean data combined for ZFAT). Although 41% (19 of 46) of the loci were nominally replicated, a higher proportion of the tested loci seemed to overlap between the Japanese and Europeans; 83% (38 of 46) of the loci showed a concordant direction of association between the two ethnic groups. Despite the substantial inter-population overlap observed in the present study, further investigation is warranted to search for loci with enhanced genetic impacts in East Asians than in Europeans.
Similar content being viewed by others
Main
Adult height is one of the most heritable human complex traits, with its heritability being estimated between 80–90%.1 Recently, several genome-wide association (GWA) studies have identified a total of 50 independent loci affecting adult height in European-descent populations.2, 3, 4, 5, 6, 7, 8, 9 Most of them, however, have turned out to exert relatively modest effects on the trait, collectively accounting for little more than 5% of height's heritability.1
Anthropometric measures such as height are known to considerably differ between ethnic groups. Thus far, inconsistent results of GWA studies have been reported in East Asian populations.10, 11 For the loci showing convincing evidence of height association in Europeans, no substantial overlap was found in the GWA study involving 618 Chinese subjects,10 whereas significant association was detected at a number of previously reported loci (that is, 14 independent loci showed P<0.05) in the GWA study involving 8842 Korean subjects.11
Along these lines, we performed the present study to test the potential overlap of height association between East Asians and Europeans and to replicate the suggestive association signals reported by the two previous GWA studies in East Asians10, 11 using the genotype data on 1530 Japanese samples, which are part of our ongoing GWA study of cardiometabolic disorders among the Japanese (Supplementary Table 1). Genotyping, data cleaning and statistical analysis in the GWA scans were performed as described elsewhere.12 The absence of population stratification was validated in the tested GWA study samples; the λ value for the genomic control was 1.04.
We examined association signals at those loci regarding which suggestive or significant association was previously reported by GWA studies in Europeans and/or East Asians2, 3, 4, 5, 6, 7, 8, 9, 10, 11 (Supplementary Tables 2 and 3). Here, single-nucleotide polymorphisms (SNPs) were tested for height association, using linear regression analysis in the additive model after adjustment for sex and age groups (Supplementary Table 4 and Supplementary Figure 1). A P-value of <0.05 (one-tailed test) was considered statistically significant when the direction of association was consistent with the original claims. We used PLINK software (http://pngu.mgh.harvard.edu/~purcell/plink/)13 and the R software package for testing the association (http://www.r-project.org). To validate modest associations detected in the GWA study samples for loci (SOCS5, ZFAT and NUP37-IGF1) with previously reported suggestive association and to test borderline associations for loci (ADAMTSL3 and HHIP) robustly replicated by the GWA studies of Europeans, we further genotyped, using TaqMan SNP Genotyping Assays (Applied Biosystems Japan, Tokyo, Japan), five SNPs in a population-based panel of 5695 Japanese described elsewhere14 (Supplementary Tables 1 and 5). The genotype distribution of all tested SNPs was in Hardy–Weinberg equilibrium (P>0.01), and the genotyping call rates were >99.5%. To assess the proportion of variance explained by an SNP, we calculated the coefficient of determination R2 as 2f(1–f)β2, where f is a minor allele frequency and β is a per-allele effect on the sex- and age-adjusted standardized height. We also performed a meta-analysis of Japanese and Korean association data and compared them with European data, using the rmeta package for the R software.
A total of 19 independent loci were found to be significantly associated with height in the Japanese population (Table 1); 18 (out of 19) loci were originally identified in Europeans, while the remaining one locus (ZFAT) was reported in Koreans. Apart from four SNPs—rs314277, rs11633371, rs7209435 and rs6088792—the effect allele frequencies were common (minor allele frequency >0.1) in the Japanese. Among the 46 loci previously reported in Europeans, although not always attaining nominal significance, 38 (83%) appeared to be similarly associated with height in the Japanese (Supplementary Table 6).
For the inter-ethnic comparison of East Asians with Europeans, we first combined our data with the results of Koreans (Supplementary Table 6). In most cases, the effect sizes of associations were relatively modest, and the explained variance (R2) fell predominantly in the range between 0.0005 and 0.003 in both ethnic groups (Figure 1 and Supplementary Figure 2). As for 21 SNPs meta-analyzed in East Asians, the direction of association was consistent with that in Europeans at all loci but one, SCMH1 (Supplementary Table 6). Also, 17% (8 out of 46) of the tested loci showed significant (P<0.05) heterogeneity in the association between East Asians and Europeans. It is therefore possible that the lack of nominally significant association at some loci in the Japanese population results from the limited sample size and/or the potential presence of ethnic heterogeneity.
Inter-ethnic comparison of per-allele effect on height between the East Asians and Europeans for 46 loci previously reported in GWA studies of Europeans and tested in the Japanese population. In the East Asians, black squares (which are proportional to the tested sample size) and vertical lines (which represent 95% CI) show the results for SNPs that are meta-analyzed with Korean data; those in gray show the results for SNPs that are analyzed solely based on the Japanese data. Curves from the top to the bottom correspond to R2=0.003, 0.002, 0.001 and 0.0005. SNP associations appear to cluster around the curve for R2=0.001 in the Europeans. On the other hand, they are widely distributed in the East Asians, for whom the curve for R2=0.0005 centers the distribution, and overlaps with the vertical lines, that is, 95% CI, for most of the 46 SNP loci.
It has to be noted that the participants are relatively old in our GWA study sample (the mean values being 66.4 and 66.0 years for males and females, respectively; Supplementary Table 1) and aging-related changes in skeletal structure, for example, osteoporosis in females, may result in decreased height among some subjects. In addition, an increasing secular trend of height in the Japanese (Supplementary Table 4 and Supplementary Figure 1) remains to be investigated extensively, although the present study tested height association at five loci in two panels, between which marked differences in age and height existed, and showed almost concordant association irrespective of the secular trend (Supplementary Table S5).
Our study could replicate significant association at a new locus, ZFAT, which has not been reported in Europeans but is likely to exert some genetic impacts on height in East Asians (Japanese and Koreans) (Table 1 and Supplementary Table 5). Apart from the genetic susceptibility to autoimmune thyroiditis reported in the Japanese,15 little is known about the function of ZFAT. Further investigation of this locus is warranted to provide novel insights into human growth.
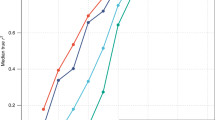
Together, the 19 associated loci (shown in Table 1) explain 6% of the height variation in the Japanese, with the effects per allele of the variants being estimated at 0.46 cm (95% confidence interval, 0.36–0.56) on average. Individuals with ⩽10 height-increasing alleles and ⩾20 height-increasing alleles differ in height by ∼6 cm (Figure 2). These figures are almost compatible with those reported to date in Europeans,5, 6, 7 despite the possibility of some over- or under-estimation owing to the limited sample size. Thus, although the effect sizes at individual loci have been proven to be small, there is substantial overlap of common variants associated with height between East Asians and Europeans.
Analysis of combined effects. For each participant in the GWA study panel of the Japanese with complete genotype at the 19 SNPs with P<0.05 in Table 1 (N=1389), we counted the number of height-increasing alleles to create a height score (x-axis). Individuals with ⩽10 or ⩾20 ‘tall’ alleles were grouped. For each height score group, the mean ±95% CI is plotted. The regression line in black indicates that each additional ‘tall’ allele increases the height by 0.46 cm. The y-axis on the right is the scale for approximate height difference from the population average. The light gray histogram in the background represents the percentage of samples in each height score group (y-axis on the left); ∼5% each in the height score ⩽10 and in the height score ⩾20.
References
Maher, B. Personal genomes: the case of the missing heritability. Nature 456, 18–21 (2008).
Weedon, M. N. & Frayling, T. M. Reaching new heights: insights into the genetics of human stature. Trends Genet. 24, 595–603 (2008).
Weedon, M. N., Lettre, G., Freathy, R. M., Lindgren, C. M., Voight, B. F., Perry, J. R. et al. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat. Genet. 39, 1245–1250 (2007).
Sanna, S., Jackson, A. U., Nagaraja, R., Willer, C. J., Chen, W. M., Bonnycastle, L. L. et al. Common variants in the GDF5-UQCC region are associated with variation in human height. Nat. Genet. 40, 198–203 (2008).
Gudbjartsson, D. F., Walters, G. B., Thorleifsson, G., Stefansson, H., Halldorsson, B. V., Zusmanovich, P. et al. Many sequence variants affecting diversity of adult human height. Nat. Genet. 40, 609–615 (2008).
Weedon, M. N., Lango, H., Lindgren, C. M., Wallace, C., Evans, D. M., Mangino, M. et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat. Genet. 40, 575–583 (2008).
Lettre, G., Jackson, A. U., Gieger, C., Schumacher, F. R., Berndt, S. I., Sanna, S. et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat. Genet. 40, 584–591 (2008).
Johansson, A., Marroni, F., Hayward, C., Franklin, C. S., Kirichenko, A. V., Jonasson, I. et al. Common variants in the JAZF1 gene associated with height identified by linkage and genome-wide association analysis. Hum. Mol. Genet. 18, 373–380 (2009).
Soranzo, N., Rivadeneira, F., Chinappen-Horsley, U., Malkina, I., Richards, J. B., Hammond, N. et al. Meta-analysis of genome-wide scans for human adult stature identifies novel loci and associations with measures of skeletal frame size. PLoS Genet. 5, e1000445 (2009).
Lei, S. F., Yang, T. L., Tan, L. J., Chen, X. D., Guo, Y., Guo, Y. F. et al. Genome-wide association scan for stature in Chinese: evidence for ethnic specific loci. Hum Genet. 125, 1–9 (2009).
Cho, Y. S., Go, M. J., Kim, Y. J., Heo, J. Y., Oh, J. H., Ban, H. J. et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat. Genet. 41, 527–534 (2009).
Takeuchi, F., Serizawa, M., Yamamoto, K., Fujisawa, T., Nakashima, E., Ohnaka, K. et al. Confirmation of multiple risk loci and genetic impacts by a genome-wide association study of type 2 diabetes in the Japanese population. Diabetes 258, 1690–1699 (2009).
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A., Bender, D. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Tsuchihashi-Makaya, M., Serizawa, M., Yanai, K., Katsuya, T., Takeuchi, F., Fujioka, A. et al. Gene-environmental interaction regarding alcohol-metabolizing enzymes in the Japanese general population. Hypertens. Res. 32, 207–213 (2009).
Shirasawa, S., Harada, H., Furugaki, K., Akamizu, T., Ishikawa, N., Ito, K. et al. SNPs in the promoter of a B cell-specific antisense transcript, SAS-ZFAT, determine susceptibility to autoimmune thyroid disease. Hum. Mol. Genet. 13, 2221–2231 (2004).
Acknowledgements
This work was supported by the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation Organization (NIBIO), and the International Medical Research Grant. We acknowledge the outstanding contributions of the International Medical Center of Japan (IMCJ) employees who provided technical and infrastructural support for this work. Above all, we thank the study subjects who made this work possible and who gave it value. We thank all the people who continuously support the Hospital-based Cohort Study in IMCJ and the Amagasaki Study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website (http://www.nature.com/jhg)
Supplementary information
Rights and permissions
About this article
Cite this article
Takeuchi, F., Nabika, T., Isono, M. et al. Evaluation of genetic loci influencing adult height in the Japanese population. J Hum Genet 54, 749–752 (2009). https://doi.org/10.1038/jhg.2009.99
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2009.99
Keywords
This article is cited by
-
Patterns of enrichment and acceleration in evolutionary rates of promoters suggest a role of regulatory regions in cetacean gigantism
BMC Ecology and Evolution (2023)
-
Analysis of a large dataset reveals haplotypes carrying putatively recessive lethal and semi-lethal alleles with pleiotropic effects on economically important traits in beef cattle
Genetics Selection Evolution (2019)
-
Reaffirmation of known major genes and the identification of novel candidate genes associated with carcass-related metrics based on whole genome sequence within a large multi-breed cattle population
BMC Genomics (2019)
-
Comprehensive genome and transcriptome analyses reveal genetic relationship, selection signature, and transcriptome landscape of small-sized Korean native Jeju horse
Scientific Reports (2019)
-
Recent progress in the study of the genetics of height
Human Genetics (2011)