Abstract
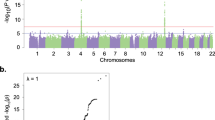
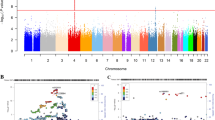
We report a GWAS of alcohol dependence (AD) in European-American (EA) and African-American (AA) populations, with replication in independent samples of EAs, AAs and Germans. Our sample for discovery and replication was 16 087 subjects, the largest sample for AD GWAS to date. Numerous genome-wide significant (GWS) associations were identified, many novel. Most associations were population specific, but in several cases were GWS in EAs and AAs for different SNPs at the same locus,showing biological convergence across populations. We confirmed well-known risk loci mapped to alcohol-metabolizing enzyme genes, notably ADH1B (EAs: Arg48His, P=1.17 × 10−31; AAs: Arg369Cys, P=6.33 × 10−17) and ADH1C in AAs (Thr151Thr, P=4.94 × 10−10), and identified novel risk loci mapping to the ADH gene cluster on chromosome 4 and extending centromerically beyond it to include GWS associations at LOC100507053 in AAs (P=2.63 × 10−11), PDLIM5 in EAs (P=2.01 × 10−8), and METAP in AAs (P=3.35 × 10−8). We also identified a novel GWS association (1.17 × 10−10) mapped to chromosome 2 at rs1437396, between MTIF2 and CCDC88A, across all of the EA and AA cohorts, with supportive gene expression evidence, and population-specific GWS for markers on chromosomes 5, 9 and 19. Several of the novel associations implicate direct involvement of, or interaction with, genes previously identified as schizophrenia risk loci. Confirmation of known AD risk loci supports the overall validity of the study; the novel loci are worthy of genetic and biological follow-up. The findings support a convergence of risk genes (but not necessarily risk alleles) between populations, and, to a lesser extent, between psychiatric traits.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Gelernter J, Sherva R, Koesterer R, Almasy L, Zhao H, Kranzler HR et al. Genomewide association study of cocaine dependence and related traits: FAM53B identified as a risk gene. Mol Psychiatry, advance online publication, 20 August 2013; doi:10.1038/mp.2013.99 (e-pub ahead of print).
Gelernter J, Kranzler HR, Sherva R, Koesterer R, Almasy L, Zhao H et al. Genomewide association study of opioid dependence: multiple associations mapped to calcium and potassium pathways. Biological Psychiatry, (in press).
Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature 2008; 452: 638–642.
Gelernter J, Kranzler HR . Genetics of alcohol dependence. Hum Genet 2009; 1: 91–99.
Li D, Zhao H, Gelernter J . Strong association of the alcohol dehydrogenase 1B gene (ADH1B) with alcohol dependence and alcohol-induced medical diseases. Biol Psychiatry 2011; 6: 504–512.
Li D, Zhao H, Gelernter J . Further clarification of the contribution of the ADH1C gene to vulnerability of alcoholism and selected liver diseases. Hum Genet 2012a; 131: 1361–1374.
Luo X, Kranzler HR, Zuo L, Yang BZ, Lappalainen J, Gelernter J . ADH4 gene variation is associated with alcohol and drug dependence in European Americans: results from family-controlled and population-structured association studies. Pharmacogenet Genomics 2005; 15: 755–768.
Li D, Zhao H, Gelernter J . Strong protective effect of the aldehyde dehydrogenase gene (ALDH2) 504lys (*2) allele against alcoholism and alcohol-induced medical diseases in Asians. Hum Genet 2012b; 131: 725–737.
Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P et al. Genome-wide association study of alcohol dependence. Arch Gen Psychiatry 2009; 66: 773–784.
Schumann G, Coin LJ, Lourdusamy A, Charoen P, Berger KH, Stacey D et al. Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc Natl Acad Sci USA 2011; 108: 7119–7124.
Frank J, Cichon S, Treutlein J, Ridinger M, Mattheisen M, Hoffmann P et al. Genome-wide significant association between alcohol dependence and a variant in the ADH gene cluster. Addict Biol 2012; 17: 171–180.
Zuo L, Wang K, Zhang XY, Krystal JH, Li CS, Zhang F et al. NKAIN1-SERINC2 is a functional, replicable and genome-wide significant risk gene region specific for alcohol dependence in subjects of European descent. Drug Alcohol Depend 2013; 129: 254–264.
Park BL, Kim JW, Cheong HS, Kim LH, Lee BC, Seo CH et al. Extended genetic effects of ADH cluster genes on the risk of alcohol dependence: from GWAS to replication. Hum Genet 2013; 132: 657–668.
Quillen EE, Chen X-D, Almasy L, Yang F, He H, Li X et alGWAS of alcohol dependence and related traits in an isolated rural Chinese sample, submitted.
Wang JC, Foroud T, Hinrichs AL, Le NX, Bertelsen S, Budde JP et al. A genome-wide association study of alcohol-dependence symptom counts in extended pedigrees identifies C15orf53. Mol Psychiatry 2012 doi:10.1038/mp.2012.143.
Pierucci-Lagha A, Gelernter J, Feinn R, Cubells JF, Pearson D, Pollastri A et al. Diagnostic reliability of the semi-structured assessment for drug dependence and alcoholism (SSADDA). Drug Alcohol Depend 2005; 80: 303–312.
American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders 4th Ed. American Psychiatric Press: Washington, DC, USA, 1994.
Holland P M, Abramson RD, Watson R, Gelfand DH . Detection of specific polymerase chain reaction product by utilizing the 5‘→3‘ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA 1991; 88: 7276–7280.
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D . Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006; 38: 904–909.
Patterson N, Price AL, Reich D . Population structure and eigen analysis. PLoS Genet 2006; 2: e190.
Zeger SL, Liang KY . Longitudinal data analysis for discrete and continuous outcomes. Biometrics 1986; 42: 121–130.
Willer CJ, Li Y, GR Abecasis . METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010; 26: 2190–2191.
Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD . Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci 2012; 32: 1884–1897.
Bierut LJ, Goate AM, Breslau N, Johnson EO, Bertelsen S, Fox L et al. ADH1B is associated with alcohol dependence and alcohol consumption in populations of European and African ancestry. Mol Psychiatry 2012; 17: 445–450.
Edenberg H J, Xuei X, Chen H-J, Tian H, Wetherill L F, Dick DM et al. Association of alcohol dehydrogenase genes with alcohol dependence: a comprehensive analysis. Hum Mol Genet 2006; 15: 1539–1549.
Horiuchi Y, Arai M, Niizato K, Iritani S, Noguchi E, Ohtsuki T et al. A polymorphism in the PDLIM5 gene associated with gene expression and schizophrenia. Biol Psychiatry 2006; 59: 434–439.
Li C, Tao R, Qin W, Zheng Y, He G, Shi Y et al. Positive association between PDLIM5 and schizophrenia in the Chinese Han population. Int J Neuropsychopharmacol 2008; 11: 27–34.
Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E et al. Gene, environment association studies consortium: a genome-wide association study of alcohol dependence. Proc Natl Acad Sci USA 2010; 107: 5082–5087.
Enomoto A, Asai N, Namba T, Wang Y, Kato T, Tanaka M et al. Roles of disrupted-in-schizophrenia 1-interacting protein girdin in postnatal development of the dentate gyrus. Neuron 2009; 63: 774–787.
Kim JY, Liu CY, Zhang F, Duan X, Wen Z, Song J et al. Interplay between DISC1 and GABA signaling regulates neurogenesis in mice and risk for schizophrenia. Cell 2012; 148: 1051.
Xie P, Kranzler HR, Krystal JH, Farrer LA, Zhao H, Gelernter J . Deep resequencing of 17 glutamate system genes identifies rare variants in DISC1 and GRIN2B affecting risk of opioid dependence. Addict Biol, advance online publication, 16 July 2013; doi:10.1111/adb.12072 (e-pub ahead of print).
Kitamura T, Asai N, Enomoto A, Maeda K, Kato T, Ishida M et al. Regulation of VEGF-mediated angiogenesis by the Akt/PKB substrate Girdin. Nat Cell Biol 2008; 10: 329–337.
Warner-Schmidt JL, Duman RS . VEGF is an essential mediator of the neurogenic and behavioral actions of antidepressants. Proc Natl Acad Sci USA 2007; 104: 4647–4652.
Blumberg HP, Wang F, Chepenik LG, Kalmar JH, Edmiston E, Duman RS et al. Influence of vascular endothelial growth factor variation on human hippocampus morphology. Biol Psychiatry 2008; 15: 901–903.
Dick DM, Meyers J, Aliev F, Nurnberger J Jr, Kramer J, Kuperman S et al. Evidence for genes on chromosome 2 contributing to alcohol dependence with conduct disorder and suicide attempts. Am J Med Genet B Neuropsychiatr Genet 2010; 153B: 1179–1188.
Chowdhury S, Chen Y, Yao TW, Ajami K, Wang XM, Popov Y et al. Regulation of dipeptidyl peptidase 8 and 9 expression in activated lymphocytes and injured liver. World J Gastroenterol 2013; 19: 2883–2893.
St Clair D, Blackwood D, Muir W, Carothers A, Walker M, Spowart G et al. Association within a family of a balanced autosomal translocation with major mental illness. Lancet 1990; 336: 13–16.
Kim JY, Duan X, Liu CY, Jang M-H, Guo JU, Pow-anpongkul N et al. DISC1 regulates new neuron development in the adult brain via modulation of AKT-mTOR signaling through KIAA1212. Neuron 2009; 63: 761–773.
Thomson PA, Parla JS, McRae AF, Kramer M, Ramakrishnan K, Yao J et al. 708 Common and 2010 rare DISC1 locus variants identified in 1542 subjects: analysis for association with psychiatric disorder and cognitive traits. Mol Psychiatry 2013 doi:10.1038/mp.2013.68.
Acknowledgements
We appreciate the work in recruitment and assessment provided at Yale University School of Medicine and the APT Foundation by James Poling, PhD; at McLean Hospital by Roger Weiss, M.D., at the Medical University of South Carolina by Kathleen Brady, MD, Ph.D. and at the University of Pennsylvania by David Oslin, MD. Genotyping services for a part of our GWAS study were provided by the Center for Inherited Disease Research (CIDR) and Yale University (Center for Genome Analysis). CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University (contract number N01-HG-65403). We are grateful to Ann Marie Lacobelle, Catherine Aldi and Christa Robinson for their excellent technical assistance, to the SSADDA interviewers, led by Yari Nuñez and Michelle Slivinsky, who devoted substantial time and effort to phenotype the study sample and to John Farrell and Alexan Mardigan for database management assistance. This study was supported by National Institutes of Health grants RC2 DA028909, R01 DA12690, R01 DA12849, R01 DA18432, R01 AA11330, R01 AA017535 and the VA Connecticut and Philadelphia VA MIRECCs. The publicly available data sets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000092.v1.p1 through dbGaP accession number phs000092.v1.p. Funding support for the Study of Addiction: Genetics and Environment (SAGE) was provided through the NIH Genes, Environment and Health Initiative [GEI] (U01 HG004422). SAGE is one of the genome-wide association studies funded as part of the Gene Environment Association Studies (GENEVA) under GEI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for collection of data sets and samples was provided by the Collaborative Study on the Genetics of Alcoholism (COGA; U10 AA008401), the Collaborative Genetic Study of Nicotine Dependence (COGEND; P01 CA089392) and the Family Study of Cocaine Dependence (FSCD; R01 DA013423). Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH GEI (U01HG004438), the National Institute on Alcohol Abuse and Alcoholism, the National Institute on Drug Abuse and the NIH contract ‘High throughput genotyping for studying the genetic contributions to human disease’ (HHSN268200782096C).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Although unrelated to the current study, Dr Kranzler has been a consultant or advisory board member for Alkermes, Lilly, Lundbeck, Pfizer and Roche. He is also a member of the American Society of Clinical Psychopharmacology's Alcohol Clinical Trials Initiative, which is supported by Lilly, Lundbeck, AbbVie and Pfizer.
Additional information
Supplementary Information accompanies the paper on the Molecular Psychiatry website
PowerPoint slides
Rights and permissions
About this article
Cite this article
Gelernter, J., Kranzler, H., Sherva, R. et al. Genome-wide association study of alcohol dependence:significant findings in African- and European-Americans including novel risk loci. Mol Psychiatry 19, 41–49 (2014). https://doi.org/10.1038/mp.2013.145
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mp.2013.145
Keywords
This article is cited by
-
Individual Trajectories of Depressive Symptoms Within Racially-Ethnically Diverse Youth: Associations with Polygenic Risk for Depression and Substance Use Intent and Perceived Harm
Behavior Genetics (2024)
-
Polygenic Effects on Individual Rule Breaking, Peer Rule Breaking, and Alcohol Sips Across Early Adolescence in the ABCD Study
Research on Child and Adolescent Psychopathology (2023)
-
Identifying alcohol misuse biotypes from neural connectivity markers and concurrent genetic associations
Translational Psychiatry (2022)
-
Genome-wide meta-analysis of alcohol use disorder in East Asians
Neuropsychopharmacology (2022)
-
Gene-based polygenic risk scores analysis of alcohol use disorder in African Americans
Translational Psychiatry (2022)