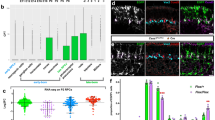
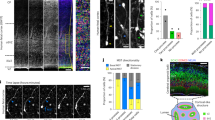
Abstract
Neurons in most regions of the mammalian nervous system are generated over an extended period of time during development. Maintaining sufficient numbers of progenitors over the course of neurogenesis is essential to ensure that neural cells are produced in correct numbers and diverse types1,2,3. The underlying molecular mechanisms, like those governing stem-cell self-renewal in general, remain poorly understood. We report here that mouse numb and numblike (Nbl)4,5,6, two highly conserved homologues of Drosophila numb7,8, play redundant but critical roles in maintaining neural progenitor cells during embryogenesis, by allowing their progenies to choose progenitor over neuronal fates. In Nbl mutant embryos also conditionally mutant for mouse numb in the nervous system, early neurons emerge in the expected spatial and temporal pattern, but at the expense of progenitor cells, leading to a nearly complete depletion of dividing cells shortly after the onset of neurogenesis. Our findings show that a shared molecular mechanism, with mouse Numb and Nbl as key components, governs the self-renewal of all neural progenitor cells, regardless of their lineage or regional identities.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Anderson, D. A. Stem cells and pattern formation in the nervous system: the possible versus the actual. Neuron 30, 19–35 (2001)
Edlund, T. & Jessell, T. M. Progression from extrinsic to intrinsic signaling in cell fate specification: a view from the nervous system. Cell 96, 211–224 (1999)
McConnell, S. K. Constructing the cerebral cortex: neurogenesis and fate determination. Neuron 15, 761–768 (1995)
Zhong, W., Feder, J. N., Jiang, M.-M., Jan, L. Y. & Jan, Y. N. Asymmetric localization of a mammalian Numb homolog during mouse cortical neurogenesis. Neuron 17, 43–53 (1996)
Zhong, W., Jiang, M.-M., Weinmaster, G., Jan, L. Y. & Jan, Y. N. Differential expression of mammalian Numb, Numblike and Notch1 suggests distinct roles during mouse cortical neurogenesis. Development 124, 1887–1897 (1997)
Verdi, J. M. et al. Mammalian NUMB is an evolutionarily conserved signaling adapter protein that specifies cell fate. Curr. Biol. 6, 113–145 (1996)
Rhyu, M. S., Jan, L. Y. & Jan, Y. N. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell 76, 477–491 (1994)
Spana, E. P., Kopczynski, C., Goodman, C. S. & Doe, C. Q. Asymmetric localization of numb autonomously determines sibling neuron identity in the Drosophila CNS. Development 121, 3489–3494 (1995)
Wakamatsu, Y., Maynard, T. M., Jones, S. U. & Weston, J. A. NUMB localizes in the basal cortex of mitotic avian neuroepithelial cells and modulates neuronal differentiation by binding to NOTCH-1. Neuron 23, 71–81 (1999)
Zilian, O. et al. Multiple roles of mouse numb in tuning developmental cell fates. Curr. Biol. 11, 494–501 (2001)
Zhong, W. et al. Mouse numb is an essential gene involved in cortical neurogenesis. Proc. Natl Acad. Sci. USA 97, 6844–6849 (2000)
Gu, M., Marth, J. D., Orban, P. C., Mossmann, H. & Rajewsky, K. Deletion of a DNA polymerase β gene segment in T cells using cell type-specific gene targeting. Science 265, 103–106 (1994)
Zimmerman, L. et al. Independent regulatory elements in the nestin gene direct transgene expression to neural stem cells or muscle precursors. Neuron 12, 11–24 (1994)
Soriano, P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nature Genet. 21, 70–71 (1999)
Dunwoodie, S. L., Henrique, D., Harrison, S. M. & Beddington, R. S. P. Mouse Dll3: a novel divergent Delta gene which may complement the function of other Delta homologues during early pattern formation in the mouse embryo. Development 124, 3065–3076 (1997)
Campos, L. S., Duarte, A. J., Branco, T. & Herique, D. mDll1 and mDll3 expression in the developing mouse brain: role in the establishment of the early cortex. J. Neurosci. Res. 64, 590–598 (2001)
Ohtsuka, T. et al. Hes1 and Hes5 as Notch effectors in mammalian neuronal differentiation. EMBO J. 18, 2196–2207 (1999)
Kuhlbrodt, K., Herbarth, B., Sock, E., Hermans-Borgmeyer, I. & Wegner, M. Sox10, a novel transcriptional modulator in glial cells. J. Neurosci. 18, 237–250 (1998)
Lu, Q. R. et al. Sonic hedgehog-regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron 25, 317–329 (2000)
Zhou, Q., Wang, S. & Anderson, D. J. Identification of a novel family of oligodendrocyte lineage-specific basic helix-loop-helix transcription factors. Neuron 25, 331–343 (2000)
Takebayashi, H. et al. Dynamic expression of basic helix-loop-helix Olig family members: implication of Olig2 in neuron and oligodendrocyte differentiation and identification of a new member, Olig3. Mech. Dev. 99, 143–148 (2000)
Mizuguchi, R. et al. Combinatorial roles of olig2 and neurogenin2 in the coordinated induction of pan-neuronal and subtype-specific properties of motoneurons. Neuron 31, 757–771 (2001)
Novitch, B. G., Chen, A. I. & Jessell, T. M. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron 31, 773–789 (2001)
Pfaff, S. L., Mendelsohn, M., Stewart, C. L., Edlund, T. & Jessell, T. M. Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell 84, 309–320 (1996)
Chenn, A. & McConnell, S. K. Cleavage orientation and the asymmetric inheritance of Notch1 immunoreactivity in mammalian neurogenesis. Cell 82, 631–641 (1995)
Qian, X., Goderie, S. K., Shen, Q., Stern, J. H. & Temple, S. Intrinsic programs of patterned cell lineages in isolated vertebrate CNS ventricular zone cells. Development 125, 3143–3152 (1998)
Noctor, S. C., Flint, A. C., Weissman, T. A., Dammerman, R. S. & Kriegstein, A. R. Neurons derived from radial glial cells establish radial units in neocortex. Nature 409, 714–720 (2001)
Cayouette, M., Whitmore, A. V., Jeffery, G. & Raff, M. Asymmetric segregation of Numb in retinal development and the influence of the pigmented epithelium. J. Neurosci. 21, 5643–5651 (2001)
Zhong, W., Mirkovitch, J. & Darnell, J. E. Jr Tissue specific regulation of mouse hepatocyte nuclear factor 4 expression. Mol. Cell. Biol. 14, 7276–7284 (1994)
Acknowledgements
We thank members of the Zhong laboratory for discussions, C. Jacobs and H. Keshishian for comments, K. Jakobsdottir for support (P.H.P.), J. Zhang for technical assistance, Developmental Studies Hybridoma Bank, D. Anderson, R. Kageyama, U. Lendahl, M. Lewandoski, M. Nakafuku, H. Takebayashi and M. Wegner for reagents. K.Z. is supported by a Yale Anderson fellowship. Y.N.J. is a HHMI investigator. This work was supported by a Yale Hellman Family fellowship and grants from March of Dimes and NINDS to W.Z. and from NIMH to the Silvio Conte Center for Neuroscience Research at UCSF.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Rights and permissions
About this article
Cite this article
Petersen, P., Zou, K., Hwang, J. et al. Progenitor cell maintenance requires numb and numblike during mouse neurogenesis. Nature 419, 929–934 (2002). https://doi.org/10.1038/nature01124
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature01124
This article is cited by
-
Therapeutic Effectiveness of Anticancer Agents Targeting Different Signaling Molecules Involved in Asymmetric Division of Cancer Stem Cell
Stem Cell Reviews and Reports (2023)
-
Dysregulation of Notch-FGF signaling axis in germ cells results in cystic dilation of the rete testis in mice
Journal of Cell Communication and Signaling (2022)
-
KIF20A/MKLP2 regulates the division modes of neural progenitor cells during cortical development
Nature Communications (2018)
-
Nestin-expressing progenitor cells: function, identity and therapeutic implications
Cellular and Molecular Life Sciences (2018)
-
Molecular components and polarity of radial glial cells during cerebral cortex development
Cellular and Molecular Life Sciences (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.