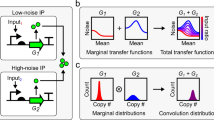
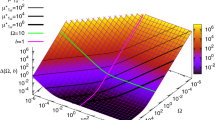
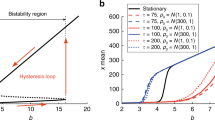
Abstract
Transcription in eukaryotic cells has been described as quantal1, with pulses of messenger RNA produced in a probabilistic manner2,3. This description reflects the inherently stochastic nature4,5,6,7,8,9 of gene expression, known to be a major factor in the heterogeneous response of individual cells within a clonal population to an inducing stimulus10,11,12,13,14,15,16. Here we show in Saccharomyces cerevisiae that stochasticity (noise) arising from transcription contributes significantly to the level of heterogeneity within a eukaryotic clonal population, in contrast to observations in prokaryotes15, and that such noise can be modulated at the translational level. We use a stochastic model of transcription initiation specific to eukaryotes to show that pulsatile mRNA production, through reinitiation, is crucial for the dependence of noise on transcriptional efficiency, highlighting a key difference between eukaryotic and prokaryotic sources of noise. Furthermore, we explore the propagation of noise in a gene cascade network and demonstrate experimentally that increased noise in the transcription of a regulatory protein leads to increased cell–cell variability in the target gene output, resulting in prolonged bistable expression states. This result has implications for the role of noise in phenotypic variation and cellular differentiation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hume, D. A. Probability in transcriptional regulation and its implications for leukocyte differentiation and inducible gene expression. Blood 96, 2323–2328 (2000)
Ross, I. L., Browne, C. M. & Hume, D. A. Transcription of individual genes in eukaryotic cells occurs randomly and infrequently. Immunol. Cell Biol. 72, 177–185 (1994)
Walters, M. C. et al. Enhancers increase the probability but not the level of gene expression. Proc. Natl Acad. Sci. USA 92, 7125–7129 (1995)
Ko, M. S. H. A stochastic model for gene induction. J. Theor. Biol. 153, 181–194 (1991)
McAdams, H. H. & Arkin, A. Stochastic mechanisms in gene expression. Proc. Natl Acad. Sci. USA 94, 814–819 (1997)
Hasty, J., Pradines, J., Dolnik, M. & Collins, J. J. Noise-based switches and amplifiers for gene expression. Proc. Natl Acad. Sci. USA 97, 2075–2080 (2000)
Thattai, M. & van Oudenaarden, A. Intrinsic noise in gene regulatory networks. Proc. Natl Acad. Sci. USA 98, 8614–8619 (2001)
Kepler, T. B. & Elston, T. C. Stochasticity in transcriptional regulation: origins, consequences, and mathematical representations. Biophys. J. 81, 3116–3136 (2001)
Swain, P. S., Elowitz, M. B. & Siggia, E. D. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc. Natl Acad. Sci. USA 99, 12795–12800 (2002)
Novick, A. & Weiner, M. Enzyme induction as an all-or-none phenomenon. Proc. Natl Acad. Sci. USA 43, 553–566 (1957)
Ko, M. S. H., Nakauchi, H. & Takahashi, N. The dose dependence of glucocorticoid-inducible gene expression results from changes in the number of transcriptionally active templates. EMBO J. 9, 2835–2842 (1990)
Fiering, S. et al. Single cell assay of a transcription factor reveals a threshold in transcription activated by signals emanating from the T-cell antigen receptor. Genes Dev. 4, 1823–1834 (1990)
van Roon, M. A., Aten, J. A., van Oven, C. H., Charles, R. & Lamers, W. H. The initiation of hepatocyte-specific gene expression within embryonic hepatocytes is a stochastic event. Dev. Biol. 136, 508–516 (1989)
Becskei, A., Séraphin, B. & Serrano, L. Positive feedback in eukaryotic gene networks: cell differentiation by graded to binary response conversion. EMBO J. 20, 2528–2535 (2001)
Ozbudak, E. M., Thattai, M., Kurtser, I., Grossman, A. D. & van Oudenaarden, A. Regulation of noise in the expression of a single gene. Nature Genet. 31, 69–73 (2002)
Elowitz, M. B., Levine, A. J., Siggia, E. D. & Swain, P. S. Stochastic gene expression in a single cell. Science 297, 1183–1186 (2002)
Ptashne, M. & Gann, A. Transcriptional activation by recruitment. Nature 386, 569–577 (1997)
Bhaumik, S. R. & Green, M. R. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 15, 1935–1945 (2001)
Larschan, E. & Winston, F. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 15, 1946–1956 (2001)
Rossi, F. M. V., Kringstein, A. M., Spicher, A., Guicherit, O. M. & Blau, H. M. Transcriptional control: rheostat converted to on/off switch. Mol. Cell 6, 723–728 (2000)
Biggar, S. R. & Crabtree, G. R. Cell signaling can direct either binary or graded transcriptional responses. EMBO J. 20, 3167–3176 (2001)
Chatterjee, S. & Struhl, K. Connecting a promoter-bound protein to TBP bypasses the need for a transcriptional activation domain. Nature 374, 820–822 (1995)
Klages, N. & Strubin, M. Stimulation of RNA polymerase II transcription initiation by recruitment of TBP in vivo. Nature 374, 822–823 (1995)
Struhl, K. Chromatin structure and RNA polymerase II connection: implications for transcription. Cell 84, 179–182 (1996)
Hahn, S. Activation and the role of reinitiation in the control of transcription by RNA polymerase II. Cold Spring Harb. Symp. Quant. Biol. 63, 181–188 (1998)
Sharp, P. M. & Li, W. H. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15, 1281–1295 (1987)
Thattai, M. & van Oudenaarden, A. Attenuation of noise in ultrasensitive signaling cascades. Biophys. J. 82, 2943–2950 (2002)
Kemkemer, R., Schrank, S., Vogel, W., Gruler, H. & Kaufmann, D. Increased noise as an effect of haploinsufficiency of the tumour-suppressor gene neurofibromatosis type 1 in vitro. Proc. Natl Acad. Sci. USA 99, 13783–13788 (2002)
Stewart, J. J. & Stargell, L. A. The stability of the TFIIA-TBP-DNA complex is dependent on the sequence of the TATAAA element. J. Biol. Chem. 276, 30078–30084 (2001)
Yean, D. & Gralla, J. Transcription reinitiation rate: a special role for the TATA box. Mol. Cell. Biol. 17, 3809–3816 (1997)
Acknowledgements
We thank T. Gilmore for guidance and helpful advice on switch construction, F. Isaacs for helpful discussions and advice, and B. Cormack for the gift of pEGFP3. This work was supported by DARPA, NSF and the Danish Research Agency.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Blake, W., KÆrn, M., Cantor, C. et al. Noise in eukaryotic gene expression. Nature 422, 633–637 (2003). https://doi.org/10.1038/nature01546
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature01546
This article is cited by
-
An analysis of codon utilization patterns in the chloroplast genomes of three species of Coffea
BMC Genomic Data (2023)
-
Comparative and phylogenetic analyses of nine complete chloroplast genomes of Orchidaceae
Scientific Reports (2023)
-
Complete chloroplast genome sequence of Gynostemma guangxiense: genome structure, codon usage bias, and phylogenetic relationships in Gynostemma (Cucurbitaceae)
Brazilian Journal of Botany (2023)
-
Periodic synchronization of isolated network elements facilitates simulating and inferring gene regulatory networks including stochastic molecular kinetics
BMC Bioinformatics (2022)
-
Noise reduction by upstream open reading frames
Nature Plants (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.