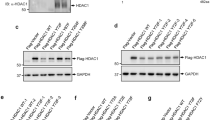
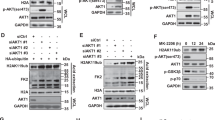
Abstract
Aberrant transcriptional repression through chromatin remodelling and histone deacetylation has been postulated to represent a driving force underlying tumorigenesis because histone deacetylase inhibitors have been found to be effective in cancer treatment. However, the molecular mechanisms by which transcriptional derepression would be linked to tumour suppression are poorly understood. Here we identify the transcriptional repressor Pokemon (encoded by the Zbtb7 gene) as a critical factor in oncogenesis. Mouse embryonic fibroblasts lacking Zbtb7 are completely refractory to oncogene-mediated cellular transformation. Conversely, Pokemon overexpression leads to overt oncogenic transformation both in vitro and in vivo in transgenic mice. Pokemon can specifically repress the transcription of the tumour suppressor gene ARF through direct binding. We find that Pokemon is aberrantly overexpressed in human cancers and that its expression levels predict biological behaviour and clinical outcome. Pokemon's critical role in cellular transformation makes it an attractive target for therapeutic intervention.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Lin, R. J. et al. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature 391, 811–814 (1998)
Melnick, A. et al. Critical residues within the BTB domain of PLZF and Bcl-6 modulate interaction with corepressors. Mol. Cell. Biol. 22, 1804–1818 (2002)
Barna, M., Hawe, N., Niswander, L. & Pandolfi, P. P. Plzf regulates limb and axial skeletal patterning. Nature Genet. 25, 166–172 (2000)
Ye, B. H. et al. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nature Genet. 16, 161–170 (1997)
Adhikary, S. et al. Miz1 is required for early embryonic development during gastrulation. Mol. Cell. Biol. 23, 7648–7657 (2003)
Carter, M. G. et al. Mice deficient in the candidate tumor suppressor gene Hic1 exhibit developmental defects of structures affected in the Miller–Dieker syndrome. Hum. Mol. Genet. 9, 413–419 (2000)
Chen, W. Y. et al. Heterozygous disruption of Hic1 predisposes mice to a gender-dependent spectrum of malignant tumors. Nature Genet. 33, 197–202 (2003)
Chen, Z. et al. Fusion between a novel Kruppel-like zinc finger gene and the retinoic acid receptor-alpha locus due to a variant t(11;17) translocation associated with acute promyelocytic leukaemia. EMBO J. 12, 1161–1167 (1993)
Ye, B. H. et al. Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large-cell lymphoma. Science 262, 747–750 (1993)
Davies, J. M. et al. Novel BTB/POZ domain zinc-finger protein, LRF, is a potential target of the LAZ-3/BCL-6 oncogene. Oncogene 18, 365–375 (1999)
Kukita, A. et al. Osteoclast-derived zinc finger (OCZF) protein with POZ domain, a possible transcriptional repressor, is involved in osteoclastogenesis. Blood 94, 1987–1997 (1999)
Pessler, F., Pendergrast, P. S. & Hernandez, N. Purification and characterization of FBI-1, a cellular factor that binds to the human immunodeficiency virus type 1 inducer of short transcripts. Mol. Cell. Biol. 17, 3786–3798 (1997)
Weinberg, R. A. The cat and mouse games that genes, viruses, and cells play. Cell 88, 573–575 (1997)
Zindy, F. et al. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 12, 2424–2433 (1998)
Serrano, M., Lin, A. W., McCurrach, M. E., Beach, D. & Lowe, S. W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88, 593–602 (1997)
Evan, G. I. et al. Induction of apoptosis in fibroblasts by c-myc protein. Cell 69, 119–128 (1992)
de Stanchina, E. et al. E1A signaling to p53 involves the p19(ARF) tumor suppressor. Genes Dev. 12,2434–2442 (1998)
Wright, W. E., Binder, M. & Funk, W. Cyclic amplification and selection of targets (CASTing) for the myogenin consensus binding site. Mol. Cell. Biol. 11, 4104–4110 (1991)
Bates, S. et al. p14ARF links the tumour suppressors RB and p53. Nature 395, 124–125 (1998)
Rowland, B. D. et al. E2F transcriptional repressor complexes are critical downstream targets of p19(ARF)/p53-induced proliferative arrest. Cancer Cell 2, 55–65 (2002)
Jacobs, J. J. L., Kieboom, K., Marino, S., DePinho, R. A. & van Lohuizen, M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 397, 164–168 (1999)
Jacobs, J. J. et al. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev. 13, 2678–2690 (1999)
Smith, K. S. et al. Bmi-1 regulation of INK4A-ARF is a downstream requirement for transformation of hematopoietic progenitors by E2a-Pbx1. Mol. Cell 12, 393–400 (2003)
Kranc, K. R. et al. Transcriptional coactivator Cited2 induces Bmi1 and Mel18 and controls fibroblast proliferation via Ink4a/ARF. Mol. Cell. Biol. 23, 7658–7666 (2003)
Kim, J. H. et al. The Bmi-1 oncoprotein is overexpressed in human colorectal cancer and correlates with the reduced p16INK4a/p14ARF proteins. Cancer Lett. 203, 217–224 (2004)
Vonlanthen, S. et al. The bmi-1 oncoprotein is differentially expressed in non-small cell lung cancer and correlates with INK4A-ARF locus expression. Br. J. Cancer 84, 1372–1376 (2001)
Jacobs, J. J. et al. Senescence bypass screen identifies TBX2, which represses Cdkn2a (p19(ARF)) and is amplified in a subset of human breast cancers. Nature Genet. 26, 291–299 (2000)
Lingbeek, M. E., Jacobs, J. J. & van Lohuizen, M. The T-box repressors TBX2 and TBX3 specifically regulate the tumor suppressor gene p14ARF via a variant T-site in the initiator. J. Biol. Chem. 277, 26120–26127 (2002)
Brummelkamp, T. R. et al. TBX-3, the gene mutated in Ulnar-Mammary Syndrome, is a negative regulator of p19ARF and inhibits senescence. J. Biol. Chem. 277, 6567–6572 (2002)
Inoue, K., Roussel, M. F. & Sherr, C. J. Induction of ARF tumor suppressor gene expression and cell cycle arrest by transcription factor DMP1. Proc. Natl Acad. Sci. USA 96, 3993–3998 (1999)
Quelle, D. E., Zindy, F., Ashmun, R. A. & Sherr, C. J. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell 83, 993–1000 (1995)
Sherr, C. J. & McCormick, F. The RB and p53 pathways in cancer. Cancer Cell 2, 103–112 (2002)
Lowe, S. W. & Sherr, C. J. Tumor suppression by Ink4a-Arf: progress and puzzles. Curr. Opin. Genet. Dev. 13, 77–83 (2003)
Dimri, G. P. et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo . Proc. Natl Acad. Sci. USA 92, 9363–9367 (1995)
Iritani, B. M., Forbush, K. A., Farrar, M. A. & Perlmutter, R. M. Control of B cell development by Ras-mediated activation of Raf. EMBO J. 16, 7019–7031 (1997)
Morse, H. C. III et al. Bethesda proposals for classification of lymphoid neoplasms in mice. Blood 100, 246–258 (2002)
Donlon, J. A., Jaffe, E. S. & Braylan, R. C. Terminal deoxynucleotidyl transferase activity in malignant lymphomas. N. Engl. J. Med. 297, 461–464 (1977)
Jaffe, E. S., Harris, N. L., Stein, H. & Vardiman, J. W. Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues (IARC Press, Lyon, France, 2001)
Whitehurst, C. E., Chattopadhyay, S. & Chen, J. Control of V(D)J recombinational accessibility of the D beta 1 gene segment at the TCR beta locus by a germline promoter. Immunity 10, 313–322 (1999)
Gurrieri, C. et al. Loss of the tumor suppressor PML in human cancers of multiple histologic origins. J. Natl Cancer Inst. 96, 269–279 (2004)
Alizadeh, A. A. et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403, 503–511 (2000)
Rosenwald, A. et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N. Engl. J. Med. 346, 1937–1947 (2002)
Hans, C. P. et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood 103, 275–282 (2004)
Shvarts, A. et al. A senescence rescue screen identifies BCL6 as an inhibitor of anti-proliferative p19ARF-p53 signaling. Genes Dev. 16, 681–686 (2002)
Lossos, I. S. et al. Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. N. Engl. J. Med. 350, 1828–1837 (2004)
Sanchez-Beato, M., Sanchez-Aguilera, A. & Piris, M. A. Cell cycle deregulation in B-cell lymphomas. Blood 101, 1220–1235 (2003)
Phan, R. T. & Dalla-Favera, R. The BCL6 proto-oncogene suppresses p53 expression in germinal-center B cells. Nature 432, 635–639 (2004)
Carnero, A., Hudson, J. D., Price, C. M. & Beach, D. H. p16INK4A and p19ARF act in overlapping pathways in cellular immortalization. Nature Cell Biol. 2, 148–155 (2000)
Hedvat, C. V. et al. Application of tissue microarray technology to the study of non-Hodgkin's and Hodgkin's lymphoma. Hum. Pathol. 33, 968–974 (2002)
Acknowledgements
We thank D. Yao, C. Hedvat, M. Dudas, J. Qin and A. Wilton for assistance on TMA preparation, staining and statistical analyses; S. Hasan for assistance with data input and management; K. Manova, C. Farrell and other Molecular Cytology Core Facility members for advice and assistance with IHC; J. Overholser and other Monoclonal Antibody Core Facility staff for help with antibody generation; G. Cattoretti and R. Dalla-Favera for advice; and L. Khandker, L. Dong, M. Hu, L. DiSantis and other P.P.P. laboratory members for assistance and discussion. This work is supported in part by an NCI grant to P.P.P.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
Supplementary Figure S1
The protein expression levels of introduced oncogenes. (JPG 25 kb)
Supplementary Figure S2
Identification of Pokemon binding sequence. (JPG 65 kb)
Supplementary Figure S3
Schematic representations of the ARF promoter. (JPG 97 kb)
Supplementary Figure S4
The expression levels of Pokemon mRNA in transgenic founder lines. (JPG 29 kb)
Supplementary Figure S4
POKEMON expression in human and mouse normal lymphoid tissues. (JPG 86 kb)
Supplementary Figure S6
Kaplan-Meier curves for various prognostic markers. (JPG 75 kb)
Supplementary Tables S1 and S2
Clinical and immunohistochemical characteristics of DLBCL patients (Supplementary Table S1), and Clinical and immunohistochemical characteristics of FL patients (Supplementary Table S2). (DOC 44 kb)
Supplementary Figure Legends
Legends to accompany the above Supplementary Figures. (DOC 58 kb)
Supplementary Methods
Contains details of additional methods (retrovirus infection; real-time PCR analysis; ChIP assay; pokemon mRNA expression level in transgenic founder lines; flow cytometry analysis; immunohistochemistry for paraffin-embedded tissues; and the statistical analysis) used in this study, and an additional reference. (DOC 42 kb)
Rights and permissions
About this article
Cite this article
Maeda, T., Hobbs, R., Merghoub, T. et al. Role of the proto-oncogene Pokemon in cellular transformation and ARF repression. Nature 433, 278–285 (2005). https://doi.org/10.1038/nature03203
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature03203
This article is cited by
-
ZBTB7A suppresses glioblastoma tumorigenesis through the transcriptional repression of EPB41L5
Experimental & Molecular Medicine (2023)
-
ZBTB7A, a miR-144-3p targeted gene, accelerates bladder cancer progression via downregulating HIC1 expression
Cancer Cell International (2022)
-
Impact of SNPs, off-targets, and passive permeability on efficacy of BCL6 degrading drugs assigned by virtual screening and 3D-QSAR approach
Scientific Reports (2022)
-
The transcription factor LRF promotes integrin β7 expression by and gut homing of CD8αα+ intraepithelial lymphocyte precursors
Nature Immunology (2022)
-
Elevated ZBTB7A expression in the tumor invasive front correlates with more tumor budding formation in gastric adenocarcinoma
Journal of Cancer Research and Clinical Oncology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.