Abstract
Development of immature T-cell precursors (thymocytes) to either the CD4 helper or CD8 killer T-cell lineages correlates precisely with their T-cell receptor specificity for major histocompatibility complex class II or class I molecules, respectively, indicating that the process is carefully regulated. Although intensively studied owing to its importance in determining the composition of the mature T-cell compartment and as a general model of binary lineage decisions, the underlying molecular pathways remain obscure. We have previously reported a spontaneous mouse mutant (HD (helper deficient) mice) in which lineage commitment is specifically perturbed without affecting positive selection. Here we show that a point mutation in the zinc finger transcription factor Th-POK (T-helper-inducing POZ/Krüppel-like factor) is responsible for redirection of class-II-restricted thymocytes to the CD8 lineage in HD mice. Furthermore, we demonstrate that constitutive expression of this factor during thymic development leads to redirection of class-I-restricted thymocytes to the CD4 lineage, indicating that Th-POK is a master regulator of lineage commitment.
Similar content being viewed by others
Main
Developing αβ T cells progress through three major stages in the thymus, defined by differential expression of the CD4 and CD8 co-receptor molecules; that is, CD4- CD8- (double negative), CD4+ CD8+ (double positive) and CD4+ CD8- or CD4- CD8+ (single positive). The double-positive to single-positive transition depends on productive rearrangement of both α- and β-subunits of the T-cell receptor (TCR) and engagement of the complete αβ TCR by intrathymic ligands (positive selection). Simultaneously, thymocytes diverge into the functionally distinct T-helper and T-killer lineages, defined by expression of CD4 and CD8, respectively. Mature T cells show an almost perfect correlation between CD4 or CD8 expression and their TCR specificity towards class II or class I major histocompatibility complex (MHC) molecules, respectively. Alternative instructive and stochastic/selective models have been proposed to explain this marked correlation (for recent reviews see refs 1, 2). Current thinking favours a quantitative version of the instructive model, whereby lineage choice is determined by the relative strength or duration of TCR engagement3,4,5,6,7,8,9; however, the intracellular pathways that are involved remain unknown.
Progress in the field has been hindered because lineage commitment is so intimately tied to the process of positive selection that it is difficult to study in isolation. Hence, no specific pathways have been identified that are required for lineage commitment but not positive selection. Recently, we identified a spontaneous recessive mutation in mice, the HD mutation, which appeared to identify a genetic locus specifically required for lineage commitment10. Notably, this mutation caused redirection of all class-II-restricted thymocytes to the CD8 lineage11. The existence of such a mutation demonstrated a mechanistic distinction between the pathways governing lineage commitment and positive selection, and provided a uniquely informative tool for dissecting the intracellular pathways that govern lineage commitment. Here we identify the HD locus as the zinc finger transcription factor Th-POK. We show that its expression in the thymus is normally restricted to the CD4 lineage, and that constitutive expression leads to redirection of class-I-restricted thymocytes to the CD4 lineage.
No primary defect in CD4 or CD8 expression in HD mice
One possible basis for lineage redirection in HD-/- mice is a primary defect in expression of one of the co-receptor molecules. For instance, in CD4-/- mice a substantial proportion of class-II-restricted thymocytes undergoes redirected development to the CD8 lineage3,12. In HD-/- mice, class-II-restricted thymocytes undergo aberrant downregulation of CD4 and upregulation of CD8, either of which could be the primary cause of altered lineage development. To test whether loss of CD4 expression is causing the defect, a constitutively expressed CD4 transgene13 was introduced onto the HD-/- background. Fluorescence-activated cell sorting (FACS) comparison of thymocytes from HD-/- and HD+/- mice expressing the CD4 transgene shows that CD4 development is not restored (Fig. 1a). HD+/- mice bearing the CD4 transgene generate mature thymocytes and peripheral T cells of both lineages, that is, single-positive CD4+ and double-positive cells (the latter corresponding to CD8-committed cells), whereas HD-/- mice generate only double-positive thymocytes (CD8 committed). To test the converse possibility, that aberrant upregulation of CD8 might be the primary cause of the HD defect, HD-/- mice were crossed to CD8a-/- mice. Significantly, HD-/- CD8a-/- double-deficient animals did not exhibit restoration of mature, single-positive CD4+ thymocytes or peripheral CD4+ T cells, indicating that aberrant CD8 upregulation is not the primary defect in HD-/- mice (Fig. 1a). A substantial population of mature double-negative T cells is observed in the thymus and periphery of these mice, presumably consisting of class-II-restricted cells that continue to develop to the CD8 lineage but cannot express surface CD8 (Fig. 1a). This analysis demonstrates that aberrant expression of CD4 or CD8 cannot by itself be responsible for altered lineage commitment of class-II-restricted thymocytes in HD-/- mice. Instead, CD4 and CD8 expression are regulated coordinately in a manner consistent with normal commitment to the CD8 lineage. In further support of normal CD8 commitment, redirected class-II-restricted thymocytes from HD-/- β2m-/- mice show upregulation of the CD8 lineage-specific gene perforin (Fig. 1b).
a, FACS analysis of thymocytes and peripheral blood lymphocytes (PBLs) from HD-/- and HD+/- mice either expressing a constitutive CD4 transgene (Tg+) or lacking CD8 expression, stained with anti-CD4, anti-CD8, anti-HSA (heat-stable antigen or CD24) and anti-CD62L. Gating on CD62L+ HSA- thymic subsets reveals fully mature thymocytes (second column), and gating on Thy1+ PBLs reveals double-negative T cells in HD-/- CD8-/- mice. At least two mice of each genotype were analysed and all showed normal thymic cellularity (0.7–2.3 × 108). Values of boxed regions are percentages. b, Real-time RT–PCR analysis of perforin mRNA levels in sorted single-positive thymocytes from indicated mice. Duplicate bars represent different mice and error bars represent standard deviations.
No detectable TCR signalling defect in HD mice
Given the important role of the TCR in lineage commitment, it is possible that the HD phenotype reflects a defect in TCR signalling. We have previously reported that several proximal events in the TCR signalling cascade including phosphorylation of CD3-ζ, ZAP-70 and p56lck are normal in HD-/- thymocytes11. We have now found that TCR-mediated induction of Ca2+ flux, and phosphorylation of the MAP kinases Erk, Jnk and p38, are also unaltered (data not shown). Positive and negative selection provide sensitive readouts of the efficiency of TCR signalling in vivo. If redirection of class-II-restricted thymocytes in HD-/- mice is due to an overall deficiency in TCR signalling, then selection defects should also be apparent for class-I-restricted TCRs. To test this, the class-I-restricted HY TCR—which recognizes a male-specific antigen and mediates positive and negative selection on female and male backgrounds, respectively—was introduced onto the HD-/- background. Positive selection was examined in RAG-/- females to preclude expression of endogenous TCR specificities. No significant difference was detected between HY+ HD-/- and HY+ HD+/+ female mice in terms of the number of single-positive CD8+ thymocytes generated, indicating that positive selection of class-I-restricted TCRs is unaffected by the HD mutation (Supplementary Fig. S1). Similarly, HY-mediated negative selection was normal in HD-/- male mice, as demonstrated by severe reductions of double-positive and single-positive thymic populations (Supplementary Fig. S1). Altogether, these data reinforce the conclusion that the HD mutation only affects lineage commitment. If the defect lies in a TCR-mediated signalling pathway, it must be a specialized pathway not required for positive or negative selection.
Genetic mapping of the HD defect
Lacking obvious candidate genes, we decided to map the HD locus genetically. For this purpose, HD-/- mice, which arose from a complex mixture of BALB/c, C57BL/6, 129/Sv and C3H strains, were crossed to wild mouse subspecies Mus musculus castaneus and Mus musculus molossinus, from which they were expected to differ at most polymorphic markers. The resulting HD+/- F1 animals were backcrossed to HD-/- mice to generate a panel of >2,000 N2 hybrid mice. All N2 animals were typed for the presence of peripheral CD4+ T cells by FACS analysis, and a subset of 635 mice (∼ 25%) was identified that exhibited the HD phenotype. Because the HD mutation is recessive, these animals were by definition homozygous for the mutant HD allele (HD-/-). By typing a fraction of these HD-/- N2 animals for a panel of chromosome-specific markers, the HD locus was mapped to mouse chromosome 3 (Fig. 2a). Narrower resolution was obtained by typing all 635 N2 HD-/- animals for a panel of closely spaced chromosome-3-specific markers. Finally, analysis of the 20 most informative recombinant mice narrowed the location of the HD mutation to an interval of <1 cM between markers D3Mit49 and D3Mit341 (Fig. 2b). On the basis of genetic mapping data for the mouse and genomic sequence data for the syntenic human region available at the time, about 30 genes could be tentatively identified as mapping within the HD region. Polymerase chain reaction (PCR) analysis of HD-/- genomic DNA proved positive for all of these genes, ruling out a major genomic deletion.
a, Genetic linkage analysis demonstrating a high degree of co-segregation of the HD allele with markers on mouse chromosome 3. b, Fine mapping of the HD locus using closely spaced chromosome 3 markers on a subset of highly informative recombinant mice. Heterozygous or homozygous typing outcomes are designated by A/B and B/B, respectively (the A/A haplotype is impossible in this breeding scheme). No data (ND) indicates instances when polymorphic markers do not distinguish between BALB/c and either M. m. molossinus or M. m. castaneus haplotypes. Question marks indicate ambiguous typing results. The c and m prefixes in the third column indicate M. m. castaneus and M. m. molossinus, respectively. Numbers in the genotype column refer to the relevant MIT marker (for example, 73 indicates D3Mit73).
Identification of the HD locus
To map the mutation more precisely, we used a bacterial artificial chromosome (BAC) transgenic complementation approach using a set of eight BAC clones, which collectively spanned the entire genetically defined HD region, as defined by D3Mit341 and D3Mit49 markers (Fig. 3a). BAC transgenic mice were crossed to HD-/- mice to generate BAC+ HD-/- N2 animals. On the basis of the restoration of single-positive CD4 cell numbers in the thymus and periphery, two of seven BACs (A126P10 and A368D24) were able to correct the HD phenotype (Fig. 3a; see also Supplementary Table S1 and Supplementary Fig. S2). Not all lines that carried these two BACs mediated phenotypic rescue, probably due to fragmentation of large BAC clones during DNA preparation or injection. The ∼160-kilobase (kb) region of overlap between these two BACs, which must logically contain the HD locus, included ten known intact genes (Fig. 3a).
a, Map of genetically defined HD region with full-length BAC clones superimposed. Grey shading indicates clones that show rescue. b, Rescue by truncated BAC subclones (internal deletions indicated by thin lines). Grey shading indicates clones that show rescue. The minimum HD region as defined by full-length BAC and BAC subclone rescue is indicated in each panel as ‘HD region’.
To confirm and refine the BAC complementation analysis, a series of truncated BAC derivatives was generated from the rescuing BAC clones and again used for transgenic complementation (Fig. 3b). Eight deletion variants rescued the HD phenotype, and all of them shared a common region of 33 kb (‘HD region’ in Fig. 3b). This region contained two intact genes: Lep503, which encodes a protein of unknown function and is expressed in epithelial cells, and Th-POK (also known as cKrox, Zfp67), a transcription factor (Figs 3b and 4)14. Clones 368.6 and 368.28, which did not complement the HD phenotype, contained intact Lep503 but a truncated Th-POK gene, resulting in loss of the last 47 amino acids (Figs 3b and 4a), implying that Th-POK is the HD gene.
a, b, Diagram of Th-POK gene organization (a) and protein domain structure (b). Location of the HD mutation is indicated by an asterisk. Breakpoint of the non-rescuing BAC subclone 368.6/28 is indicated as ‘ΔBAC’. c, Nucleotide and amino acid sequence of the second zinc finger domain showing the location of the HD mutation.
To confirm this, we searched for mutations in Th-POK transcripts from HD-/- mice. Complementary DNAs from HD mutant mice showed a specific A to G change at nucleotide position 1165 within the Th-POK coding region, resulting in an Arg to Gly substitution at amino acid position 389 (Fig. 4c). This mutation was confirmed in multiple, independently cloned cDNAs as well as in genomic DNA from HD mice. All wild-type Th-POK sequences determined by us or found in public databases encode an Arg at this position, including mouse sequences from BALB/c, C57BL/6, FVB and CZECH II strains, as well as homologues from human and five other vertebrate species, implying that this residue is functionally critical. The Arg to Gly substitution occurs within the second of four zinc finger domains of Th-POK (Fig. 4b), and affects a residue predicted to interact directly with DNA (according to the model of ref. 15). It has been shown that mutagenesis of key individual residues within zinc finger domains can abolish their DNA binding ability16, suggesting that the HD defect results from failure of Th-POK to bind to a key regulatory site(s).
Th-POK is expressed specifically in the CD4 lineage
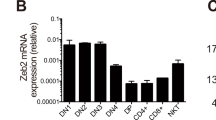
On the basis of our previous demonstration that the HD defect is intrinsic to the T-cell lineage11, we predicted that Th-POK should be expressed in developing thymocytes. Initial northern blot analysis confirmed that Th-POK RNA is expressed in the thymus as well as many other tissues (Supplementary Fig. S3), consistent with a previous report14 and data from expressed sequence tag (EST) databases. Real-time PCR with reverse transcription (RT–PCR) analysis of sorted wild-type thymic subsets revealed a marked stage- and lineage-specific expression pattern, such that Th-POK transcripts were restricted to mature single-positive CD4+ and intermediate CD4+ CD8lo subsets (Fig. 5). The latter subset contains precursors of both the CD4 and CD8 lineages. To distinguish between class-I- and class-II-restricted cells at this stage, we used mice in which only T cells of one or the other specificity can be selected; that is, mice deficient for either class I or class II MHC or expressing a particular TCR transgene. Notably, Th-POK messenger RNA expression was preferentially upregulated in class-II-restricted CD4+ CD8lo cells, consistent with a critical early role in lineage specification (Fig. 5). Interestingly, in HD-/- β2m-/- mice, in which all selected cells are class-II-restricted, Th-POK is upregulated at the CD4+ CD8lo stage but returns to background levels in mature single-positive CD8 thymocytes. This correlation between loss of CD4 expression and Th-POK downregulation suggests that Th-POK expression is controlled by TCR-mediated signalling.
Th-POK expression as measured by real-time RT–PCR analysis is shown for sorted thymocyte subsets from indicated mice. Samples from mice in which only class-I- or class-II-restricted thymocytes can be positively selected are indicated by black or white bars, respectively, whereas those from mice with a normal TCR repertoire are indicated in grey. Duplicate bars for sorted subsets of the same genotype represent different mice. I0, MHC class-I-deficient; II0, MHC class-II-deficient; DN, double negative; DP, double positive. AND and OT-1 are MHC class-II- and class-I-restricted TCR transgenes, respectively. Error bars represent standard deviations.
Ectopic Th-POK drives CD4 development
The lineage-specific expression pattern of Th-POK suggested that its induction might be sufficient for CD4 commitment. In this case, enforced constitutive expression of Th-POK should direct all thymocytes to the CD4 lineage. To test this, we used two complementary approaches: retroviral transduction of bone marrow and stable transgenesis using T-cell specific promoters. The retroviral approach allows very high transgene expression in the haematopoietic compartment and identification of cells expressing the transgene due to an associated green fluorescent protein (GFP) marker. Wild-type and mutant (Arg389Gly) Th-POK cDNAs in the MigR1 retroviral vector17 were transduced into HD+/+ or HD-/- bone marrow and then transferred into RAG-/- recipients, which lack endogenous T cells. After reconstitution of the haematopoietic compartment, FACS analysis was carried out on GFP+ lymphocytes from thymus and peripheral blood. Notably, overexpression of wild-type Th-POK mediated exclusive generation of single-positive CD4+ thymocytes and peripheral T cells in both HD-/- and wild-type backgrounds (Fig. 6a). Single-positive CD4+ thymocytes exhibited a normal proportion of mature TCRhi CD69lo cells. The fact that almost all single-positive CD4+ thymocytes were TCRhi indicates that they underwent normal positive selection, which requires TCR expression and engagement, and implies that Th-POK overexpression cannot mediate development to the single-positive CD4+ stage without positive selection. In contrast, mutant Th-POK had no effect on thymic development in either wild-type or HD-/- backgrounds. The ability of the wild-type but not mutant forms of Th-POK to correct the HD defect demonstrates that the Arg389Gly mutation, rather than another uncharacterized mutation linked to the Th-POK locus, is responsible for the HD defect. The inability of mutant Th-POK to shift development of wild-type thymocytes towards the CD8 lineage, even when expressed at very high levels under the control of retroviral enhancers, argues strongly that the mutant form is functionally inert and unable to compete with the wild-type form.
a, FACS analysis of GFP+ thymocytes and PBLs from RAG-/- mice, 5 weeks after reconstitution with wild-type or HD-/- bone marrow transduced with either wild-type or mutant Th-POK retroviral constructs, as indicated. b, FACS analysis of thymocytes and PBLs from HD-/- or HD+/- mice expressing a wild-type Th-POK transgene. SP, single positive. c, FACS analysis of thymocytes and PBLs from OT-1 transgenic or MHC class-II-deficient (II0) mice in the presence or absence of a Th-POK transgene. Thymocytes were stained with anti-CD4, anti-CD8, anti-TCR-β and anti-CD69, and PBLs with anti-CD4 and anti-CD8 antibodies. TCR and CD69 expression is shown for gated CD4+ CD8- thymocytes in the second rows of a and b. CD4 and CD8 expression is shown for gated, mature TCR+ CD69- thymocytes in the third row of b and second row of c. OT-1 PBLs were additionally stained with anti-Vα2 and anti-Vβ5 antibodies in c. At least two animals of each genotype were analysed with similar results. The number at the upper right of each plot indicates total thymic cellularity. d, Real-time RT–PCR analysis of perforin and Gata3 expression in sorted, single-positive CD4 CD69+ and CD69- thymocyte subsets from Th-POK transgenic OT-1 or MHC class-II-deficient mice (black bars), or equivalent control mice (white bars). Error bars represent standard deviations.
The above experiments show that constitutive Th-POK expression prevents development of single-positive CD8+ thymocytes; however, it is not clear whether this effect is mediated during thymic development or earlier in haematopoiesis. In addition, the proportion of GFP+ thymocytes and the levels of GFP they express are substantially lower for wild-type compared with mutant Th-POK constructs (data not shown), suggesting that premature or excessive expression of wild-type Th-POK could partly block T-cell development. To restrict Th-POK transgene expression to the T-cell lineage and to more physiological levels, we generated stable transgenic lines in which wild-type Th-POK was controlled by CD2 regulatory elements ((WT)Th-POKpCD2)18. This transgene closely reproduced the key results obtained from retroviral transduction; that is, it restored development of CD4+ T cells in an HD-/- background and abolished single-positive CD8+ cells in the thymus and periphery of both HD-/- and HD+/- backgrounds (Fig. 6b). However, the (WT)Th-POKpCD2 transgene did not cause any change in thymic cellularity, indicating that the level of Th-POK expression supported by the CD2 vector does not impair T-cell development. Single-positive CD4+ thymocytes were generated in normal numbers, and contained a normal proportion of mature TCRhi CD69lo cells. Similar results were obtained for several independent transgenic lines, in which Th-POK expression was controlled either by CD2 or CD4 regulatory elements (data not shown).
To determine whether the absence of single-positive CD8+ cells in these mice reflected redirection of class-I-restricted thymocytes to the CD4 lineage, the (WT)Th-POKpCD2 transgene was crossed to a class-I-restricted TCR transgene, OT-1. Notably, single-positive CD8+ thymocytes and peripheral T cells were entirely replaced by single-positive CD4+ cells, consistent with redirection (Fig. 6c). In the periphery, most single-positive CD4+ cells expressed both transgenic Vα2 and Vβ5 chains, indicating that they had been selected on the basis of the OT-1 TCR (Fig. 6c). To obtain independent confirmation of this result, a different wild-type Th-POK transgenic line ((WT)Th-POKpCD4) was introduced onto a MHC class-II-deficient background, thereby again limiting the selected TCR repertoire to class-I-restricted specificities. In these mice, only mature single-positive CD4+ cells were generated in the thymus and periphery in contrast to non-transgenic MHC class-II-deficient control mice, which produced only single-positive CD8 cells (Fig. 6c). The absolute number of peripheral CD4+ T cells in OT-1+ and MHC class-II-deficient mice expressing Th-POK transgenes was lower than that of CD8+ T cells in non-transgenic control mice, indicating a reduced efficiency in thymic emigration and/or peripheral survival of class-I-restricted cells lacking CD8. A selective disadvantage for OT-1+ cells lacking CD8 would be expected based on studies in OT-1+ CD8-/- mice19. Finally, to demonstrate by criteria other than co-receptor expression pattern that redirected class-I-restricted thymocytes in Th-POK transgenic mice are fully committed to the CD4 lineage, we determined RNA levels for perforin and Gata3, which represent markers of the CD8 and CD4 lineages, respectively. Notably, single-positive CD4+ cells from Th-POK transgenic mice showed absence of perforin expression and upregulation of Gata3, as expected for CD4-committed cells (Fig. 6d). The 3–5-fold elevation in Gata3 expression observed in Th-POK transgenic CD4+ cells relative to non-transgenic CD8+ cells from control mice closely matches the fourfold expression difference previously reported between normal single-positive CD4+ and CD8+ thymocytes20.
Discussion
Our results show that the presence of functional Th-POK in thymocytes undergoing positive selection is necessary and sufficient for commitment to the CD4 lineage, whereas its absence is necessary and sufficient for commitment to the CD8 lineage. The fact that Th-POK expression is confined to cells expressing a class-II-restricted TCR specificity as well as CD4 suggests that it is induced by a strong TCR signal, consistent with currently favoured quantitative instructive models of lineage commitment. Full maturation of redirected thymocytes in HD and Th-POK transgenic mice also supports an instructive model, albeit indirectly, because such maturation is incompatible with the alternative stochastic/selective model. The fact that Th-POK is not involved in other TCR-mediated processes (positive and negative selection) would suggest that it lies on a specialized branch of the TCR signalling pathway.
On the basis of the cellular distribution and structure of the HD product, we have proposed the new designation ‘T-helper-inducing POZ/Krüppel-like factor’ (Th-POK) for this locus (previously called cKrox, cKrox-α or Zfp67). Th-POK was first described as an in vitro interacting factor and transcriptional repressor of collagen genes (thus ‘collagen’ Krox or cKrox)14,21. It belongs to a group of factors related to the Drosophila segmentation protein Krüppel, and including Sp1, Egr-1, WT-1 and LKLF. Within this category, Th-POK belongs to a subset containing an amino-terminal BTP/POZ domain, the so-called POK (POZ and Krüppel) proteins, also including Bcl-6 and PLZF. The BTB/POZ domain is responsible for homo- and heterodimerization, transcriptional repression activity and interactions with components of the histone deacetylase complex, thereby providing a functional link to enzymatic activities that regulate chromatin conformation. Th-POK is highly conserved between mice and humans (100% identity for the zinc finger region). There are precedents for the critical role of zinc finger proteins in regulating haematopoietic lineage branch points, specifically Gata1 in the case of erythrocyte development22,23 and Gata3 in the Th1 versus Th2 branchpoint24. Th-POK, like Gata3, is widely expressed, and on this basis might be expected to mediate important functions in other tissues. We are currently generating a Th-POK knockout mouse to test whether it encodes any additional non-redundant functions.
Recently, two nuclear factors have been reported to be important effectors of CD4 lineage development. Gata3 is preferentially expressed in the CD4 lineage20,25 and is required for CD4 development26, whereas Runx3 is critical for CD4 silencing27. However, inactivation of either of these genes fails to cause redirection of class-II-restricted cells to the CD8 lineage26,28, suggesting that they do not regulate the commitment process itself. Furthermore, transgenic overexpression of the nuclear high-mobility group (HMG) box protein TOX mediates development of thymocytes exclusively to the CD8 lineage29. However, this occurs only in the absence of TCR engagement, not when thymocytes receive normal TCR signals, arguing against a physiological role in lineage commitment29,30.
The identification of Th-POK as a central regulator of lineage commitment provides an important new starting point for analysing the underlying molecular pathways. Future research into the transcriptional control of Th-POK itself and the nature of its target genes should contribute to unravelling these pathways further.
Methods
Mice
CD4 (ref. 13) (provided by S. Reiner) and HY TCR31 transgenic lines, as well as HD-/- mice10,11, have been previously described. M. m. castaneus and M. m. molossinus strains were purchased from Jackson Laboratories. All other transgenic lines were generated by the FCCC Transgenic Facility.
Flow cytometry
Cells were prepared from thymus and peripheral blood, and analysed by flow cytometry according to standard procedures. All antibodies were obtained from BD Pharmingen.
Real-time RT–PCR
Real-time RT–PCR analysis for Th-POK, perforin and Gata3 was carried out according to the probe-based method and analysed by the comparative Ct method (compared to β-actin). Primer and probe sequences are available upon request.
Genetic mapping
HD-/- mice, which arose from mixed C57BL/6, BALB/c, C3H and 129/Sv parentage, were crossed to M. m. castaneus or M. m. molossinus strains, and the resulting HD+/- N1 animals were again crossed to HD-/- mice, resulting in >2,000 N2 hybrid mice. A total of 635 of these mice were identified as HD-/- and used for PCR-based SSLP linkage analysis (see Supplementary Information).
Genomic sequencing
Sequencing was carried out for three C57BL/6 BAC clones (RPCI-23 295A4, 418C12 and 368D24), which collectively span most of the HD region, confirming the overlap of these three clones, presence of the D3Mit49 marker within 368D24, and presence of many of the genes predicted from BAC end sequencing and genetic data (GenBank accession numbers AC104329, AC104327 and AC104632).
BAC complementation
BAC clones spanning the HD region were identified using the BAC assembly database for the C57BL/6 RPCI-23 BAC library (Genome Sequence Centre, BC Cancer Agency). See Supplementary Information for details.
Transgenic production
BAC clones used for generation of transgenic mice were obtained from the BAC Resource Consortium. Subclones of BACs 126P10 and 368D24 bearing internal deletions were generated by restriction endonuclease treatment followed by re-ligation. The boundaries of all deletions were mapped by PCR analysis and DNA sequencing. For transgenic expression of wild-type Th-POK cDNA the full-length cDNA insert was cloned into the hCD2 vector18 or into a vector containing CD4 promoter/enhancer elements (derived from transgenic construct h in ref. 32).
Retroviral transduction
Complete wild-type and mutant Th-POK cDNAs were cloned into the BglII site of the MigR1 vector (gift of W. Pear). Preparation of viral supernatant and transduction of bone marrow was performed essentially as described17.
References
Germain, R. N. T-cell development and the CD4–CD8 lineage decision. Nature Rev. Immunol. 2, 309–322 (2002)
Bosselut, R. & Singer, A. CD4/CD8 coreceptors in thymocyte development, selection, and lineage commitment: analysis of the CD4/CD8 lineage decision. Adv. Immunol. 83, 91–131 (2004)
Matechak, E. O., Killeen, N., Hedrick, S. M. & Fowlkes, B. J. MHC class II-specific T cells can develop in the CD8 lineage when CD4 is absent. Immunity 4, 337–347 (1996)
Itano, A. et al. The cytoplasmic domain of CD4 promotes the development of CD4 lineage T cells. J. Exp. Med. 183, 731–741 (1996)
Yasutomo, K., Doyle, C., Miele, L., Fuchs, C. & Germain, R. N. The duration of antigen receptor signalling determines CD4 + versus CD8 + T-cell lineage fate. Nature 404, 506–510 (2000)
Liu, X. & Bosselut, R. Duration of TCR signaling controls CD4–CD8 lineage differentiation in vivo. Nature Immunol. 5, 280–288 (2004)
Hernandez-Hoyos, G., Sohn, S. J., Rothenberg, E. V. & Alberola-Ila, J. Lck activity controls CD4/CD8 T cell lineage commitment. Immunity 12, 313–322 (2000)
Legname, G. et al. Inducible expression of a p56Lck transgene reveals a central role for Lck in the differentiation of CD4 SP thymocytes. Immunity 12, 537–546 (2000)
Schmedt, C. et al. Csk controls antigen receptor-mediated development and selection of T-lineage cells. Nature 394, 901–904 (1998)
Davé, V. P., Allman, D., Keefe, R., Hardy, R. R. & Kappes, D. J. HD mice: a novel mouse mutant with a specific defect in the generation of CD4(+ ) T cells. Proc. Natl Acad. Sci. USA 95, 8187–8192 (1998)
Keefe, R., Dave, V., Allman, D., Wiest, D. & Kappes, D. J. Regulation of lineage commitment distinct from positive selection. Science 286, 1149–1153 (1999)
Tyznik, A. J., Sun, J. C. & Bevan, M. J. The CD8 population in CD4-deficient mice is heavily contaminated with MHC class II-restricted T cells. J. Exp. Med. 199, 559–565 (2004)
Killeen, N. & Littman, D. R. Helper T-cell development in the absence of CD4-p56lck association. Nature 364, 729–732 (1993)
Galera, P., Musso, M., Ducy, P. & Karsenty, G. c-Krox, a transcriptional regulator of type I collagen gene expression, is preferentially expressed in skin. Proc. Natl Acad. Sci. USA 91, 9372–9376 (1994)
Klevit, R. E. Recognition of DNA by Cys2,His2 zinc fingers. Science 253, 1367–1393 (1991)
Nardelli, J., Gibson, T. J., Vesque, C. & Charnay, P. Base sequence discrimination by zinc-finger DNA-binding domains. Nature 349, 175–178 (1991)
Pui, J. C. et al. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity 11, 299–308 (1999)
Zhumabekov, T., Corbella, P., Tolaini, M. & Kioussis, D. Improved version of a human CD2 minigene based vector for T cell-specific expression in transgenic mice. J. Immunol. Methods 185, 133–140 (1995)
Goldrath, A. W., Hogquist, K. A. & Bevan, M. J. CD8 lineage commitment in the absence of CD8. Immunity 6, 633–642 (1997)
Hernández-Hoyos, G., Anderson, M. K., Wang, C., Rothenberg, E. V. & Alberola-Ila, J. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity 19, 83–94 (2003)
Widom, R. L., Lee, J. Y., Joseph, C., Gordon-Froome, I. & Korn, J. H. The hcKrox gene family regulates multiple extracellular matrix genes. Matrix Biol. 20, 451–462 (2001)
Tsai, S. F. et al. Cloning of cDNA for the major DNA-binding protein of the erythroid lineage through expression in mammalian cells. Nature 339, 446–451 (1989)
Blobel, G. A., Simon, M. C. & Orkin, S. H. Rescue of GATA-1-deficient embryonic stem cells by heterologous GATA-binding proteins. Mol. Cell. Biol. 15, 626–633 (1995)
Zheng, W. & Flavell, R. A. The transcription factor GATA-3 is necessary and sufficient for the Th2 cytokine gene expression in CD4 T cells. Cell 89, 587–596 (1997)
Hendriks, R. W. et al. Expression of the transcription factor GATA-3 is required for the development of the earliest T cell progenitors and correlates with stages of cellular proliferation in the thymus. Eur. J. Immunol. 29, 1912–1918 (1999)
Pai, S.-Y. et al. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity 19, 863–875 (2003)
Taniuchi, I. et al. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 111, 621–633 (2002)
Woolf, E. et al. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc. Natl Acad. Sci. USA 100, 7731–7736 (2003)
Aliahmad, P. et al. TOX provides a link between calcineurin activation and CD8 lineage commitment. J. Exp. Med. 199, 1089–1099 (2004)
Wilkinson, B. et al. TOX: an HMG box protein implicated in the regulation of thymocyte selection. Nature Immunol. 3, 272–280 (2002)
Kisielow, P., Bluethmann, H., Staerz, U. D., Steinmetz, M. & von Boehmer, H. Tolerance in T cell receptor transgenic mice involves deletion of immature CD4 + CD8 + thymocytes. Nature 333, 742–746 (1988)
Sawada, S., Scarborough, J. D., Killeen, N. & Littman, D. R. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell 77, 917–929 (1994)
Acknowledgements
We acknowledge technical assistance with flow cytometry by J. Oesterling, and animal husbandry by members of the Laboratory Animal Facility. V.P.D. was responsible for initial genetic mapping of the HD mutation to a resolution of 4 cM. We thank D. Wiest for comments on the manuscript. This work was supported by National Institutes of Health Grants to D.J.K, an NIH Core Grant, and also an appropriation from the Commonwealth of Pennsylvania.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
Supplementary Figures 1-3
This file contains Supplementary Figures 1 (FACS analysis of thymocytes from male and female HY+ HD / and HD+/+ mice), 2 (FACS analysis of thymocytes and PBLs from BAC transgenic HD-/- mice demonstrating restoration of CD4 development) and 3 (northern blot analysis of Th-POK mRNA expression in various mouse tissues). (PDF 871 kb)
Supplementary Table 1
Results of BAC transgenic complementation analysis. (PDF 523 kb)
Supplementary Data
This file contains Supplementary Methods (details of genetic mapping and BAC complementation analysis), Supplementary Figure legends and Supplementary Table legend. (DOC 10 kb)
Rights and permissions
About this article
Cite this article
He, X., He, X., Dave, V. et al. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature 433, 826–833 (2005). https://doi.org/10.1038/nature03338
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature03338
This article is cited by
-
ZBTB7B is a permissive regulator of hepatocellular carcinoma initiation by repressing c-Jun expression and function
Cell Death & Disease (2024)
-
ThPOK is a critical multifaceted regulator of myeloid lineage development
Nature Immunology (2023)
-
How autoreactive thymocytes differentiate into regulatory versus effector CD4+ T cells after avoiding clonal deletion
Nature Immunology (2023)
-
Markers and makers of NKT17 cells
Experimental & Molecular Medicine (2023)
-
Unique properties of tissue-resident memory T cells in the lungs: implications for COVID-19 and other respiratory diseases
Nature Reviews Immunology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.