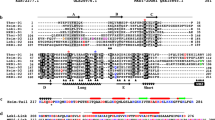
Abstract
Eukaryotic cells have two contrasting cytoskeletal and ciliary organizations. The simplest involves a single cilium-bearing centriole, nucleating a cone of individual microtubules (probably ancestral for unikonts: animals, fungi, Choanozoa and Amoebozoa). In contrast, bikonts (plants, chromists and all other protozoa) were ancestrally biciliate with a younger anterior cilium, converted every cell cycle into a dissimilar posterior cilium and multiple ciliary roots of microtubule bands. Here we show by comparative genomic analysis that this fundamental cellular dichotomy also involves different myosin molecular motors. We found 37 different protein domain combinations, often lineage-specific, and many previously unidentified. The sequence phylogeny and taxonomic distribution of myosin domain combinations identified five innovations that strongly support unikont monophyly and the primary bikont/unikont bifurcation. We conclude that the eukaryotic cenancestor (last common ancestor) had a cilium, mitochondria, pseudopodia, and myosins with three contrasting domain combinations and putative functions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Sellers, J. R. Myosins (Oxford Univ. Press, Oxford, 1999)
Bahler, M. Are class III and class IX myosins motorized signalling molecules? Biochim. Biophys. Acta 1496, 52–59 (2000)
Cavalier-Smith, T. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int. J. Syst. Evol. Microbiol. 52, 297–354 (2002)
Thompson, R. F. & Langford, G. M. Myosin superfamily evolutionary history. Anat. Rec. 268, 276–289 (2002)
Furusawa, T., Ikawa, S., Yanai, N. & Obinata, M. Isolation of a novel PDZ-containing myosin from hematopoietic supportive bone marrow stromal cell lines. Biochem. Biophys. Res. Commun. 270, 67–75 (2000)
Goodson, H. V. & Spudich, J. A. Molecular evolution of the myosin family: relationships derived from comparisons of amino acid sequences. Proc. Natl Acad. Sci. USA 90, 659–663 (1993)
Hodge, T. & Cope, M. J. A myosin family tree. J. Cell Sci. 113, 3353–3354 (2000)
Berg, J. S., Powell, B. C. & Cheney, R. E. A millennial myosin census. Mol. Biol. Cell 12, 780–794 (2001)
Kull, F. J., Vale, R. D. & Fletterick, R. J. The case for a common ancestor: kinesin and myosin motor proteins and G proteins. J. Muscle Res. Cell Motil. 19, 877–886 (1998)
Leipe, D. D., Wolf, Y. I., Koonin, E. V. & Aravind, L. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317, 41–72 (2002)
Stechmann, A. & Cavalier-Smith, T. Rooting the eukaryote tree by using a derived gene fusion. Science 297, 89–91 (2002)
Stechmann, A. & Cavalier-Smith, T. The root of the eukaryote tree pinpointed. Curr. Biol. 13, R665–R666 (2003)
Cavalier-Smith, T. Protist phylogeny and the high-level classification of Protozoa. Eur. J. Protistol. 39, 338–348 (2003)
Matsuzaki, M. et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428, 653–657 (2004)
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997)
Cavalier-Smith, T. Only six kingdoms of life. Proc. R. Soc. Lond. B 271, 1251–1262 (2004)
Simpson, A. G. & Roger, A. J. The real ‘kingdoms’ of eukaryotes. Curr. Biol. 14, R693–R696 (2004)
D'Aquino, J. A. & Ringe, D. Determinants of the SRC homology domain 3-like fold. J. Bacteriol. 185, 4081–4086 (2003)
Bapteste, E. & Philippe, H. The potential value of indels as phylogenetic markers: position of trichomonads as a case study. Mol. Biol. Evol. 19, 972–977 (2002)
Hirt, R. P. et al. Microsporidia are related to Fungi: evidence from the largest subunit of RNA polymerase II and other proteins. Proc. Natl Acad. Sci. USA 96, 580–585 (1999)
Cavalier-Smith, T. Archamoebae: the ancestral eukaryotes? BioSystems 25, 25–38 (1991)
Cavalier-Smith, T., Chao, E. E. & Oates, B. Molecular phylogeny of Amoebozoa and the evolutionary significance of the unikont Phalansterium. Eur. J. Protistol. 40, 21–48 (2004)
Bolivar, I., Fahrni, J. F., Smirnov, A. & Pawlowski, J. SSU rRNA-based phylogenetic position of the genera Amoeba and Chaos (Lobosea, Gymnamoebia): the origin of gymnamoebae revisited. Mol. Biol. Evol. 18, 2306–2314 (2001)
Kudryavtsev, A. A., Bernhardt, D., Schlegel, M., Chao, E. E. & Cavalier-Smith, T. 18S ribosomal RNA gene sequences of Cochliopodium (Himatismenida) and the phylogeny of Amoebozoa. Protist 156, 215–224 (2005)
Milyutina, I. A., Aleshin, V. V., Mikrjukov, K. A., Kedrova, O. S. & Petrov, N. B. The unusually long small subunit ribosomal RNA gene found in amitochondriate amoeboflagellate Pelomyxa palustris: its rRNA predicted secondary structure and phylogenetic implication. Gene 272, 131–139 (2001)
Dacks, J. B., Marinets, A., Doolittle, W. F., Cavalier-Smith, T. & Logsdon, J. M. Jr Analyses of RNA polymerase II genes from free-living protists: phylogeny, long branch attraction, and the eukaryotic big bang. Mol. Biol. Evol. 19, 830–840 (2002)
Bapteste, E. et al. The analysis of 100 genes supports the grouping of three highly divergent amoebae: Dictyostelium, Entamoeba, and Mastigamoeba. Proc. Natl Acad. Sci. USA 99, 1414–1419 (2002)
Lang, B. F., O'Kelly, C., Nerad, T., Gray, M. W. & Burger, G. The closest unicellular relatives of animals. Curr. Biol. 12, 1773–1778 (2002)
Berney, C., Fahrni, J. & Pawlowski, J. How many novel eukaryotic ‘kingdoms’? Pitfalls and limitations of environmental DNA surveys. BMC Biol. 2, 13 (2004)
Bass, D. et al. Polyubiquitin insertions and the phylogeny of Cercozoa and Rhizaria. Protist 156, 149–161 (2005)
Cavalier-Smith, T. & Chao, E. E. Phylogeny and classification of phylum Cercozoa (Protozoa). Protist 154, 341–358 (2003)
Titus, M. A. A class VII unconventional myosin is required for phagocytosis. Curr. Biol. 9, 1297–1303 (1999)
Tuxworth, R. I. et al. A role for myosin VII in dynamic cell adhesion. Curr. Biol. 11, 318–329 (2001)
Marchler-Bauer, A. et al. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 31, 383–387 (2003)
Bateman, A. et al. The Pfam protein families database. Nucleic Acids Res. 32, D138–D141 (2004)
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 (1997)
Ronquist, F. & Huelsenbeck, J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003)
Schmidt, H. A., Strimmer, K., Vingron, M. & von Haeseler, A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18, 502–504 (2002)
Felsenstein, J. Phylip (Department of Genetics, University of Washington, Seattle, 1995)
Minotto, L., Edwards, M. R. & Bagnara, A. S. Trichomonas vaginalis: Characterization, expression, and phylogenetic analysis of a carbamate kinase gene sequence. Exp. Parasitol. 95, 54–62 (2000)
Baldauf, S. L. & Palmer, J. D. Animals and fungi are each other's closest relatives: congruent evidence from multiple proteins. Proc. Natl Acad. Sci. USA 90, 11558–11562 (1993)
Acknowledgements
Preliminary sequence data were obtained from The Institute for Genomic Research website (http://www.tigr.org) and the Department of Energy Joint Genome Institute (JGI) website (http://www.jgi.doe.gov). We thank TIGR and DOE JGI for making data publicly available, A. A. Davies for comments and assistance with data management, and D. Soanes for PSI BLAST assistance. T.A.R. was supported by a BBSRC studentship. T.C.-S. thanks NERC for research grants and NERC and the Canadian Institute for Advanced Research Evolutionary Biology Program for Fellowship support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Table S1.
Comparative genome survey of myosin genes. Full details of comparative genomic analyses of the myosin gene family displayed as a table with footnotes and an additional table listing details about domain names, definitions and abbreviations. (DOC 185 kb)
Supplementary Figure S1.
Alignment of representatives of myosin class II, V and XI. Annotated amino acid alignment that illustrates sequence synapomorphies and details about protein domain evolution. (PDF 1803 kb)
Supplementary Figure S2.
Alignment of myosins with either MYTH4 and/or FERM domains. Annotated amino acid alignment that illustrates sequence synapomorphies and details about protein domain evolution. (PDF 2115 kb)
Supplementary Figure S3.
Alignment of representatives of myosin class I. Annotated amino acid alignment that illustrates sequence synapomorphies and details about protein domain evolution. (PDF 2638 kb)
Supplementary Figure S4.
Phylogeny of myosin head domains; this is the same tree as in Fig. 2 of the main text of the paper, except that accession numbers replace species names and exact support values are shown. The details of the phylogenetic methods are explained in the figure legend. (PDF 82 kb)
Rights and permissions
About this article
Cite this article
Richards, T., Cavalier-Smith, T. Myosin domain evolution and the primary divergence of eukaryotes. Nature 436, 1113–1118 (2005). https://doi.org/10.1038/nature03949
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature03949
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.