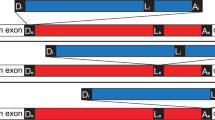
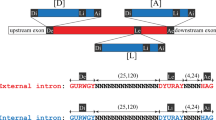
Abstract
The minor spliceosome is a ribonucleoprotein complex that catalyses the removal of an atypical class of spliceosomal introns (U12-type) from eukaryotic messenger RNAs1,2. It was first identified and characterized in animals, where it was found to contain several unique RNA constituents that share structural similarity with and seem to be functionally analogous to the small nuclear RNAs (snRNAs) contained in the major spliceosome3,4,5,6,7,8. Subsequently, minor spliceosomal components and U12-type introns have been found in plants9,10,11,12 but not in fungi. Unlike that of the major spliceosome, which arose early in the eukaryotic lineage13,14, the evolutionary history of the minor spliceosome is unclear because there is evidence of it in so few organisms. Here we report the identification of homologues of minor-spliceosome-specific proteins and snRNAs, and U12-type introns, in distantly related eukaryotic microbes (protists) and in a fungus (Rhizopus oryzae). Cumulatively, our results indicate that the minor spliceosome had an early origin: several of its characteristic constituents are present in representative organisms from all eukaryotic supergroups for which there is any substantial genome sequence information. In addition, our results reveal marked evolutionary conservation of functionally important sequence elements contained within U12-type introns and snRNAs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Patel, A. A. & Steitz, J. A. Splicing double: insights from the second spliceosome. Nature Rev. Mol. Cell Biol. 4, 960–970 (2003)
Will, C. L. & Lührmann, R. Splicing of a rare class of introns by the U12-dependent spliceosome. Biol. Chem. 386, 713–724 (2005)
Tarn, W-Y., Yario, T. A. & Steitz, J. A. U12 snRNA in vertebrates: evolutionary conservation of 5′ sequences implicated in splicing of pre-mRNAs containing a minor class of introns. RNA 1, 644–656 (1995)
Hall, S. L. & Padgett, R. A. Requirement of U12 snRNA for in vivo splicing of a minor class of eukaryotic nuclear pre-mRNA introns. Science 271, 1716–1718 (1996)
Tarn, W-Y. & Steitz, J. A. A novel spliceosome containing U11, U12, and U5 snRNPs excises a minor class (AT–AC) intron in vitro.. Cell 84, 801–811 (1996)
Tarn, W-Y. & Steitz, J. A. Highly diverged U4 and U6 small nuclear RNAs required for splicing rare AT–AC introns. Science 273, 1824–1832 (1996)
Yu, Y-T. & Steitz, J. A. Site-specific crosslinking of mammalian U11 and U6atac to the 5′ splice site of an AT–AC intron. Proc. Natl Acad. Sci. USA 94, 6030–6035 (1997)
Kolossova, I. & Padgett, R. A. U11 snRNA interacts in vivo with the 5′ splice site of U12-dependent (AU–AC) pre-mRNA introns. RNA 3, 227–233 (1997)
Burge, C. B., Padgett, R. A. & Sharp, P. A. Evolutionary fates and origins of U12-type introns. Mol. Cell 2, 773–785 (1998)
Shukla, G. C. & Padgett, R. A. Conservation of functional features of U6atac and U12 snRNAs between vertebrates and higher plants. RNA 5, 525–538 (1999)
Lorković, Z. J., Lehner, R., Forstner, C. & Barta, A. Evolutionary conservation of minor U12-type spliceosome between plants and humans. RNA 11, 1095–1107 (2005)
Zhu, W. & Brendel, V. Identification, characterization and molecular phylogeny of U12-dependent introns in the Arabidopsis thaliana genome. Nucleic Acids Res. 31, 4561–4572 (2003)
Collins, L. & Penny, D. Complex spliceosomal organization ancestral to extant eukaryotes. Mol. Biol. Evol. 22, 1053–1066 (2005)
Rodríguez-Trelles, F., Tarrío, R. & Ayala, F. J. Origin and evolution of spliceosomal introns. Annu. Rev. Genet. doi:10.1146/annurev.genet.40.110405.090625 (published online 12 June 2006).
Hall, S. L. & Padgett, R. A. Conserved sequences in a class of rare eukaryotic nuclear introns with non-consensus splice sites. J. Mol. Biol. 239, 357–365 (1994)
Dietrich, R. C., Incorvaia, R. & Padgett, R. A. Terminal intron dinucleotide sequences do not distinguish between U2- and U12-dependent introns. Mol. Cell 1, 151–160 (1997)
Anderson, I. J. et al. Gene discovery in the Acanthamoeba castellanii genome. Protist 156, 203–214 (2005)
Dsouza, M., Larsen, N. & Overbeek, R. Searching for patterns in genomic data. Trends Genet. 13, 497–498 (1997)
Levine, A. & Durbin, R. A computational scan for U12-dependent introns in the human genome sequence. Nucleic Acids Res. 29, 4006–4013 (2001)
Dietrich, R. C., Peris, M. J., Seyboldt, A. S. & Padgett, R. A. Role of the 3′ splice site in U12-dependent intron splicing. Mol. Cell. Biol. 21, 1942–1952 (2001)
Dietrich, R. C., Fuller, J. D. & Padgett, R. A. A mutational analysis of U12-dependent splice site dinucleotides. RNA 11, 1430–1440 (2005)
Will, C. L., Schneider, C., Reed, R. & Lührmann, R. Identification of both shared and distinct proteins in the major and minor spliceosomes. Science 284, 2003–2005 (1999)
Will, C. L. et al. The human 18S U11/U12 snRNP contains a set of novel proteins not found in the U2-dependent spliceosome. RNA 10, 929–941 (2004)
Schneider, C., Will, C. L., Brosius, J., Frilander, M. J. & Lührmann, R. Identification of an evolutionarily divergent U11 small nuclear ribonucleoprotein particle in Drosophila.. Proc. Natl Acad. Sci. USA 101, 9584–9589 (2004)
Otake, L. R., Scamborova, P., Hashimoto, C. & Steitz, J. A. The divergent U12-type spliceosome is required for pre-mRNA splicing and is essential for development in Drosophila.. Mol. Cell 9, 439–446 (2002)
Benecke, H., Lührmann, R. & Will, C. L. The U11/U12 snRNP 65K protein acts as a molecular bridge, binding the U12 snRNA and U11–59K protein. EMBO J. 24, 3057–3069 (2005)
Vaňáčová, S., Yan, W., Carlton, J. M. & Johnson, P. J. Spliceosomal introns in the deep-branching eukaryote Trichomonas vaginalis.. Proc. Natl Acad. Sci. USA 102, 4430–4435 (2005)
Russell, A. G., Shutt, T. E., Watkins, R. F. & Gray, M. W. An ancient spliceosomal intron in the ribosomal protein L7a gene (Rpl7a) of Giardia lamblia.. BMC Evol. Biol. 5, 45 (2005)
Russell, A. G., Schnare, M. N. & Gray, M. W. A large collection of compact box C/D snoRNAs and their isoforms in Euglena gracilis: structural, functional and evolutionary insights. J. Mol. Biol. 357, 1548–1565 (2006)
Keeling, P. J. et al. The tree of eukaryotes. Trends Ecol. Evol. 20, 670–676 (2005)
Acknowledgements
We thank M. N. Schnare, T. E. Shutt and A. J. Lohan for discussions; M. Dlutek for automated DNA sequencing; and the various genome sequencing centres for making preliminary data publicly available, including the Broad Institute of Harvard and MIT (Rhizopus) and the Joint Genome Institute (Phytophthora). This work was supported by funding (to M.W.G.) from the Canadian Institutes of Health Research and Genome Canada (Protist EST Program). M.W.G. acknowledges salary support from the Canada Research Chairs Program and the Canadian Institute for Advanced Research. Author Contributions J.M.C initially discovered the first protist minor-spliceosomal snRNA (A. castellanii U12) and A.G.R later identified the Phytophthora U11 and U12 snRNAs. A.G.R. did the intron and protein orthologue searches and experimentally verified intron removal and snRNA gene expression. D.F.S. assisted with computer analysis and isolated A. castellanii nucleic acids. M.W.G discovered the COMMD7 intron and initiated a search for minor-spliceosome-specific protein orthologues in protists. A.G.R. wrote the paper. All authors discussed the results and critiqued the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The A. castellanii U12 snRNA sequence and the additional gene copy of Aca ORF 1 are deposited in Genbank (DQ888370 and DQ888371, respectively). Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Figure 1
U12-Type Minor spliceosomal introns in Acanthamoeba castellanii. Complete annotation of the sequences, features and gene locations of the U12-type introns identified in the protist A. castellanii (PDF 38 kb)
Supplementary Figure 2
Identification of minor spliceosome-specific protein homologues. Predicted amino acid sequences and alignments of minor spliceosome-specific protein orthologues and sequence-related major spliceosomal proteins identified in this study (PDF 119 kb)
Supplementary Figure 3
U12-Type minor spliceosomal introns in Rhizopus (Fungi) and Phytophthora (Chromalveolata) Complete annotation of the sequences, features, gene locations, and evidence supporting the identification of U12-type introns in P. ramorum, P. sojae, and Rhizopus oryzae (PDF 48 kb)
Supplementary Figure 4
Secondary structures of spliceosomal U2 and U12 snRNAs and detection of Acanthamoeba castellanii U12 snRNA. Sequences and predicted secondary structures of U12 and U2 snRNAs in A. castellanii, P. ramorum, P. sojae and their human homologues. Also shown is experimental evidence for expression of A. castellanii U12 snRNA and annotation of genomic sequences encoding all the snRNAs identified in this study (PDF 183 kb)
Supplementary Figure 5
Oligonucleotide primers and strategy used to search for minor spliceosomal introns and Phytophthora U11 snRNA homologues. Contains the sequences and descriptions of oligonucleotide primers used in this study. Specific search parameters used for the identification of U12-type introns and Phytophthora U11 snRNAs are described (PDF 11 kb)
Rights and permissions
About this article
Cite this article
Russell, A., Charette, J., Spencer, D. et al. An early evolutionary origin for the minor spliceosome. Nature 443, 863–866 (2006). https://doi.org/10.1038/nature05228
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature05228
This article is cited by
-
Defective minor spliceosomes induce SMA-associated phenotypes through sensitive intron-containing neural genes in Drosophila
Nature Communications (2020)
-
Patterns of conservation of spliceosomal intron structures and spliceosome divergence in representatives of the diplomonad and parabasalid lineages
BMC Evolutionary Biology (2019)
-
Domestication of self-splicing introns during eukaryogenesis: the rise of the complex spliceosomal machinery
Biology Direct (2017)
-
On the Possibility of an Early Evolutionary Origin for the Spliced Leader Trans-Splicing
Journal of Molecular Evolution (2017)
-
Transcriptome-scale RNase-footprinting of RNA-protein complexes
Nature Biotechnology (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.