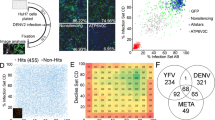
Abstract
West Nile virus (WNV), and related flaviviruses such as tick-borne encephalitis, Japanese encephalitis, yellow fever and dengue viruses, constitute a significant global human health problem1. However, our understanding of the molecular interaction of such flaviviruses with mammalian host cells is limited1. WNV encodes only 10 proteins, implying that it may use many cellular proteins for infection1. WNV enters the cytoplasm through pH-dependent endocytosis, undergoes cycles of translation and replication, assembles progeny virions in association with endoplasmic reticulum, and exits along the secretory pathway1,2,3. RNA interference (RNAi) presents a powerful forward genetics approach to dissect virus–host cell interactions4,5,6. Here we report the identification of 305 host proteins that affect WNV infection, using a human-genome-wide RNAi screen. Functional clustering of the genes revealed a complex dependence of this virus on host cell physiology, requiring a wide variety of molecules and cellular pathways for successful infection. We further demonstrate a requirement for the ubiquitin ligase CBLL1 in WNV internalization, a post-entry role for the endoplasmic-reticulum-associated degradation pathway in viral infection, and the monocarboxylic acid transporter MCT4 as a viral replication resistance factor. By extending this study to dengue virus, we show that flaviviruses have both overlapping and unique interaction strategies with host cells. This study provides a comprehensive molecular portrait of WNV–human cell interactions that forms a model for understanding single plus-stranded RNA virus infection, and reveals potential antiviral targets.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Brinton, M. A. The molecular biology of West Nile Virus: A new invader of the western hemisphere. Annu. Rev. Microbiol. 56, 371–402 (2002)
Chu, J. J. & Ng, M. L. Infectious entry of West Nile virus occurs through a clathrin-mediated endocytic pathway. J. Virol. 78, 10543–10555 (2004)
Krishnan, M. N. et al. Rab 5 is required for the cellular entry of dengue and West Nile viruses. J. Virol. 81, 4881–4885 (2007)
Brass, A. L. et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science 319, 921–926 (2008)
Ng, T. I. et al. Identification of host genes involved in hepatitis C virus replication by small interfering RNA technology. Hepatology 45, 1413–1421 (2007)
Pelkmans, L. et al. Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature 436, 78–86 (2005)
Davis, W. G., Blackwell, J. L., Shi, P. Y. & Brinton, M. A. Interaction between the cellular protein eEF1A and the 3′-terminal stem-loop of West Nile virus genomic RNA facilitates viral minus-strand RNA synthesis. J. Virol. 81, 10172–10187 (2007)
Emara, M. M. & Brinton, M. A. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc. Natl Acad. Sci. USA 104, 9041–9046 (2007)
Hirsch, A. J. et al. The Src family kinase c-Yes is required for maturation of West Nile virus particles. J. Virol. 79, 11943–11951 (2005)
Mackenzie, J. M., Khromykh, A. A. & Parton, R. G. Cholesterol manipulation by West Nile virus perturbs the cellular immune response. Cell Host Microbe 2, 229–239 (2007)
Fredericksen, B. L., Smith, M., Katze, M. G., Shi, P. Y. & Gale, M. The host response to West Nile Virus infection limits viral spread through the activation of the interferon regulatory factor 3 pathway. J. Virol. 78, 7737–7747 (2004)
Lim, J. et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell 125, 801–814 (2006)
Mashimo, T. et al. A nonsense mutation in the gene encoding 2′-5′-oligoadenylate synthetase/L1 isoform is associated with West Nile virus susceptibility in laboratory mice. Proc. Natl Acad. Sci. USA 99, 11311–11316 (2002)
Samuel, M. A. et al. PKR and RNase L contribute to protection against lethal West Nile Virus infection by controlling early viral spread in the periphery and replication in neurons. J. Virol. 80, 7009–7019 (2006)
Scherbik, S. V., Paranjape, J. M., Stockman, B. M., Silverman, R. H. & Brinton, M. A. RNase L plays a role in the antiviral response to West Nile virus. J. Virol. 80, 2987–2999 (2006)
Tecle, T., White, M. R., Gantz, D., Crouch, E. C. & Hartshorn, K. L. Human neutrophil defensins increase neutrophil uptake of influenza A virus and bacteria and modify virus-induced respiratory burst responses. J. Immunol. 178, 8046–8052 (2007)
Fujita, Y. et al. Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E-cadherin complex. Nature Cell Biol. 4, 222–231 (2002)
Dell'Angelica, E. C. et al. AP-3: An adaptor-like protein complex with ubiquitous expression. EMBO J. 16, 917–928 (1997)
Khor, R., McElroy, L. J. & Whittaker, G. R. The ubiquitin-vacuolar protein sorting system is selectively required during entry of influenza virus into host cells. Traffic 4, 857–868 (2003)
Meusser, B., Hirsch, C., Jarosch, E. & Sommer, T. ERAD: The long road to destruction. Nature Cell Biol. 7, 766–772 (2005)
Wakana, Y. et al. Bap31 is an itinerant protein that moves between the peripheral ER and a juxtanuclear compartment related to ER-associated degradation. Mol. Biol. Cell 19, 1825–1836 (2008)
Schelhaas, M. et al. Simian Virus 40 depends on ER protein folding and quality control factors for entry into host cells. Cell 131, 516–529 (2007)
Halestrap, A. P. & Price, N. T. The proton-linked monocarboxylate transporter (MCT) family: Structure, function and regulation. Biochem. J. 343, 281–299 (1999)
Chiu, M. W., Shih, H. M., Yang, T. H. & Yang, Y. L. The type 2 dengue virus envelope protein interacts with small ubiquitin-like modifier-1 (SUMO-1) conjugating enzyme 9 (Ubc9). J. Biomed. Sci. 14, 429–444 (2007)
Hanks, S. K., Ryzhova, L., Shin, N. Y. & Brabek, J. Focal adhesion kinase signaling activities and their implications in the control of cell survival and motility. Front. Biosci. 8, d982–d996 (2003)
Mi, H. et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 33, D284–D288 (2005)
Su, A. I. et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl Acad. Sci. USA 101, 6062–6067 (2004)
Mishra, G. R. et al. Human protein reference database — 2006 update. Nucleic Acids Res. 34, D411–D414 (2006)
Bader, G. D., Betel, D. & Hogue, C. W. BIND: The Biomolecular Interaction Network Database. Nucleic Acids Res. 31, 248–250 (2003)
Scholle, F. & Mason, P. W. West Nile virus replication interferes with both poly(I:C)-induced interferon gene transcription and response to interferon treatment. Virology 342, 77–87 (2005)
Rossi, S. L., Zhao, Q., O'Donnell, V. K. & Mason, P. W. Adaptation of West Nile virus replicons to cells in culture and use of replicon-bearing cells to probe antiviral action. Virology 331, 457–470 (2005)
Helenius, A., Kartenbeck, J., Simons, K. & Fries, E. On the entry of Semliki forest virus into BHK-21 cells. J. Cell Biol. 84, 404–420 (1980)
Eisen, M. B., Spellman, P. T., Brown, P. O. & Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA 95, 14863–14868 (1998)
Saldanha, A. J. Java Treeview — extensible visualization of microarray data. Bioinformatics 20, 3246–3248 (2004)
Acknowledgements
The human genome RNAi library was made available through the support of the New England Regional Center of Excellence in Biodefense and Emerging Infectious Disease (U54AI057159). The screening was performed at the ICCB-Longwood screening facility (Harvard Medical School). We thank B. Lindenbach for suggestions and Y. Benita for illustrations. We thank L2 Diagnostics for providing the anti-WNV antibody. This work was supported by the NIH. A.N. is supported by a fellowship award from the Crohn’s and Colitis Foundation of America. R.J.X. is supported by the NIH (AI062773) and by CCIB development funds. F.D.G was supported by an NIH training grant in Emerging and Tropical Infectious Diseases (AI07526); portions of this work were supported by a grant from NIAID to P.W.M. through the WRCE (NIH U54 AI057156). E.F. and S.J.E. are Investigators of the Howard Hughes Medical Institute.
Author Contributions M.N.K., H.A. and E.F. designed the experiments; M.N.K., B.S., E.F. and R.A. performed the screen; M.N.K., B.S., R.A.K., A.L.B., S.J.E. and H.A. analysed the data; M.N.K. and H.S. performed validations; P.D.U. designed microscopy; F.D.G. and P.W.M. designed replicon experiments; S.L. provided PITPNM2 cDNA; A.N. and R.J.X. performed bioinformatics analyses; and M.N.K., H.A., A.N., R.J.X. and E.F. co-wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information 1
This file contains the legends to the Supplementary Tables 1 and 2. (PDF 98 kb)
Supplementary Information 2
This file contains Supplementary Figures 1-14 and Supplementary Tables 3-9 with Legends. (PDF 19548 kb)
Supplementary Table 1
The file contains Supplementary Table 1. (XLS 185 kb)
Supplementary Table 2
The file contains Supplementary Table 2. (XLS 40 kb)
Rights and permissions
About this article
Cite this article
Krishnan, M., Ng, A., Sukumaran, B. et al. RNA interference screen for human genes associated with West Nile virus infection. Nature 455, 242–245 (2008). https://doi.org/10.1038/nature07207
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature07207
This article is cited by
-
Cdc2-like kinases: structure, biological function, and therapeutic targets for diseases
Signal Transduction and Targeted Therapy (2023)
-
Genome-wide bidirectional CRISPR screens identify mucins as host factors modulating SARS-CoV-2 infection
Nature Genetics (2022)
-
MasterPATH: network analysis of functional genomics screening data
BMC Genomics (2020)
-
The Restrictome of Flaviviruses
Virologica Sinica (2020)
-
Impaired Autophagy Flux is Associated with Proinflammatory Microglia Activation Following Japanese Encephalitis Virus Infection
Neurochemical Research (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.