Abstract
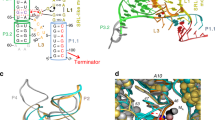
In bacteria, the intracellular concentration of several amino acids is controlled by riboswitches1,2,3,4. One of the important regulatory circuits involves lysine-specific riboswitches, which direct the biosynthesis and transport of lysine and precursors common for lysine and other amino acids1,2,3. To understand the molecular basis of amino acid recognition by riboswitches, here we present the crystal structure of the 174-nucleotide sensing domain of the Thermotoga maritima lysine riboswitch in the lysine-bound (1.9 ångström (Å)) and free (3.1 Å) states. The riboswitch features an unusual and intricate architecture, involving three-helical and two-helical bundles connected by a compact five-helical junction and stabilized by various long-range tertiary interactions. Lysine interacts with the junctional core of the riboswitch and is specifically recognized through shape-complementarity within the elongated binding pocket and through several direct and K+-mediated hydrogen bonds to its charged ends. Our structural and biochemical studies indicate preformation of the riboswitch scaffold and identify conformational changes associated with the formation of a stable lysine-bound state, which prevents alternative folding of the riboswitch and facilitates formation of downstream regulatory elements. We have also determined several structures of the riboswitch bound to different lysine analogues5, including antibiotics, in an effort to understand the ligand-binding capabilities of the lysine riboswitch and understand the nature of antibiotic resistance. Our results provide insights into a mechanism of lysine-riboswitch-dependent gene control at the molecular level, thereby contributing to continuing efforts at exploration of the pharmaceutical and biotechnological potential of riboswitches.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
Protein Data Bank
Data deposits
Atomic coordinates of the X-ray structures of the lysine riboswitch bound to ligands have been deposited in the RCSB Protein Data Bank under the following accession codes: lysine, 3DIL; AEC, 3DIG; l-4-oxalysine, 3DJ0; homoarginine, 3DIQ; and iminoethyl-l-lysine, 3DIR. Codes for other structures are: free state, 3DIS; [Ir(NH3)6]3+-soaked, 3DIO; Cs+-soaked, 3DIM; Tl+-soaked, 3DJ2; Mn2+-soaked, 3DIY; K+-anomalous, 3DIX; and Mg2+-free form, 3DIZ.
References
Sudarsan, N., Wickiser, J. K., Nakamura, S., Ebert, M. S. & Breaker, R. R. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 17, 2688–2697 (2003)
Grundy, F. J., Lehman, S. C. & Henkin, T. M. The L box regulon: lysine sensing by leader RNAs of bacterial lysine biosynthesis genes. Proc. Natl Acad. Sci. USA 100, 12057–12062 (2003)
Rodionov, D. A., Vitreschak, A. G., Mironov, A. A. & Gelfand, M. S. Regulation of lysine biosynthesis and transport genes in bacteria: yet another RNA riboswitch? Nucleic Acids Res. 31, 6748–6757 (2003)
Mandal, M. et al. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science 306, 275–279 (2004)
Blount, K. F., Wang, J. X., Lim, J., Sudarsan, N. & Breaker, R. R. Antibacterial lysine analogs that target lysine riboswitches. Nature Chem. Biol. 3, 44–49 (2007)
Serganov, A. & Patel, D. J. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nature Rev. Genet. 8, 776–790 (2007)
Nudler, E. & Mironov, A. S. The riboswitch control of bacterial metabolism. Trends Biochem. Sci. 29, 11–17 (2004)
Winkler, W. C. & Breaker, R. R. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 59, 487–517 (2005)
Chowdhury, S., Maris, C., Allain, F. H. & Narberhaus, F. Molecular basis for temperature sensing by an RNA thermometer. EMBO J. 25, 2487–2497 (2006)
Dann, C. E. et al. Structure and mechanism of a metal-sensing regulatory RNA. Cell 130, 878–892 (2007)
Klein, D. J. & Ferre-D’Amare, A. R. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science 313, 1752–1756 (2006)
Cochrane, J. C., Lipchock, S. V. & Strobel, S. A. Structural investigation of the GlmS ribozyme bound to its catalytic cofactor. Chem. Biol. 14, 97–105 (2007)
Batey, R. T., Gilbert, S. D. & Montange, R. K. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature 432, 411–415 (2004)
Serganov, A. et al. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem. Biol. 11, 1729–1741 (2004)
Serganov, A., Polonskaia, A., Phan, A. T., Breaker, R. R. & Patel, D. J. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature 441, 1167–1171 (2006)
Thore, S., Leibundgut, M. & Ban, N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science 312, 1208–1211 (2006)
Edwards, T. E. & Ferre-D’Amare, A. R. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure 14, 1459–1468 (2006)
Montange, R. K. & Batey, R. T. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature 441, 1172–1175 (2006)
Gilbert, S. D., Rambo, R. P., Van Tyne, D. & Batey, R. T. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nature Struct. Mol. Biol. 15, 177–182 (2008)
Weixlbaumer, A. et al. Crystal structure of the ribosome recycling factor bound to the ribosome. Nature Struct. Mol. Biol. 14, 733–737 (2007)
Blouin, S. & Lafontaine, D. A. A loop–loop interaction and a K-turn motif located in the lysine aptamer domain are important for the riboswitch gene regulation control. RNA 13, 1256–1267 (2007)
Correll, C. C., Freeborn, B., Moore, P. B. & Steitz, T. A. Metals, motifs, and recognition in the crystal structure of a 5S rRNA domain. Cell 91, 705–712 (1997)
Leontis, N. B. & Westhof, E. The 5S rRNA loop E: chemical probing and phylogenetic data versus crystal structure. RNA 4, 1134–1153 (1998)
Klein, D. J., Schmeing, T. M., Moore, P. B. & Steitz, T. A. The kink-turn: a new RNA secondary structure motif. EMBO J. 20, 4214–4221 (2001)
Basu, S. et al. A specific monovalent metal ion integral to the AA platform of the RNA tetraloop receptor. Nature Struct. Biol. 5, 986–992 (1998)
Feig, A. L. & Uhlenbeck, O. C. in The RNA World Second Edition (eds Gesteland, R. F., Cech, T. R. & Atkins, J. F.) 287–319 (Cold Spring Harbor Laboratory Press, 1999)
Lu, Y., Shevtchenko, T. N. & Paulus, H. Fine-structure mapping of cis-acting control sites in the lysC operon of Bacillus subtilis . FEMS Microbiol. Lett. 92, 23–27 (1992)
Patte, J. C., Akrim, M. & Mejean, V. The leader sequence of the Escherichia coli lysC gene is involved in the regulation of LysC synthesis. FEMS Microbiol. Lett. 169, 165–170 (1998)
Ataide, S. F. et al. Mechanisms of resistance to an amino acid antibiotic that targets translation. ACS Chem. Biol. 2, 819–827 (2007)
de La Fortelle, E. & Bricogne, G. in Methods in Enzymology 472–494 (Academic Press, 1997)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Perrakis, A., Morris, R. & Lamzin, V. S. Automated protein model building combined with iterative structure refinement. Nature Struct. Biol. 6, 458–463 (1999)
Acknowledgements
We thank personnel of beamline X29 at the Brookhaven National Laboratory and beamlines 24-ID-C/E at the Advanced Photon Source, Argonne National Laboratory, funded by the US Department of Energy. We thank O. Ouerfelli for the synthesis of iridium hexamine. D.J.P. was supported by funds from the National Institutes of Health.
Author Contributions L.H. crystallized the T. maritima lysine riboswitch; A.S. determined the structures and was assisted by L.H. during refinement; A.S. and L.H. performed biochemical experiments; and A.S. and D.J.P. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Supplementary Information
The file contains Supplementary Figures 1-17 and Legends, Supplementary Tables 1-3 and Supplementary Notes (PDF 8637 kb)
Rights and permissions
About this article
Cite this article
Serganov, A., Huang, L. & Patel, D. Structural insights into amino acid binding and gene control by a lysine riboswitch. Nature 455, 1263–1267 (2008). https://doi.org/10.1038/nature07326
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature07326
This article is cited by
-
Boron-assisted abiotic polypeptide synthesis
Communications Chemistry (2023)
-
Na+ riboswitches regulate genes for diverse physiological processes in bacteria
Nature Chemical Biology (2022)
-
Contact networks in RNA: a structural bioinformatics study with a new tool
Journal of Computer-Aided Molecular Design (2022)
-
Improving the Microbial Production of Amino Acids: From Conventional Approaches to Recent Trends
Biotechnology and Bioprocess Engineering (2021)
-
Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures
Nature Methods (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.