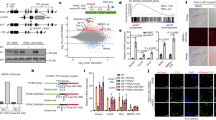
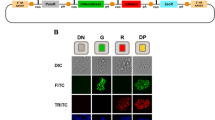
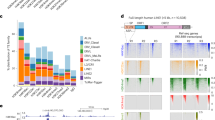
Abstract
Long interspersed element-1 (LINE-1 or L1) retrotransposition continues to affect human genome evolution1,2. L1s can retrotranspose in the germline, during early development and in select somatic cells3,4,5,6,7,8; however, the host response to L1 retrotransposition remains largely unexplored. Here we show that reporter genes introduced into the genome of various human embryonic carcinoma-derived cell lines (ECs) by L1 retrotransposition are rapidly and efficiently silenced either during or immediately after their integration. Treating ECs with histone deacetylase inhibitors rapidly reverses this silencing, and chromatin immunoprecipitation experiments revealed that reactivation of the reporter gene was correlated with changes in chromatin status at the L1 integration site. Under our assay conditions, rapid silencing was also observed when reporter genes were delivered into ECs by mouse L1s and a zebrafish LINE-2 element, but not when similar reporter genes were delivered into ECs by Moloney murine leukaemia virus or human immunodeficiency virus, suggesting that these integration events are silenced by distinct mechanisms. Finally, we demonstrate that subjecting ECs to culture conditions that promote differentiation attenuates the silencing of reporter genes delivered by L1 retrotransposition, but that differentiation, in itself, is not sufficient to reactivate previously silenced reporter genes. Thus, our data indicate that ECs differ from many differentiated cells in their ability to silence reporter genes delivered by L1 retrotransposition.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001)
Goodier, J. L. & Kazazian, H. H. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell 135, 23–35 (2008)
Garcia-Perez, J. L. et al. LINE-1 retrotransposition in human embryonic stem cells. Hum. Mol. Genet. 16, 1569–1577 (2007)
Kano, H. et al. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 23, 1303–1312 (2009)
Muotri, A. R. et al. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 435, 903–910 (2005)
Ostertag, E. M. et al. A mouse model of human L1 retrotransposition. Nature Genet. 32, 655–660 (2002)
van den Hurk, J. A. et al. L1 retrotransposition can occur early in human embryonic development. Hum. Mol. Genet. 16, 1587–1592 (2007)
Coufal, N. G. et al. L1 retrotransposition in human neural progenitor cells. Nature 460, 1127–1131 (2009)
Sperger, J. M. et al. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc. Natl Acad. Sci. USA 100, 13350–13355 (2003)
Hohjoh, H. & Singer, M. F. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 15, 630–639 (1996)
Brouha, B. et al. Evidence consistent with human L1 retrotransposition in maternal meiosis I. Am. J. Hum. Genet. 71, 327–336 (2002)
Gilbert, N., Lutz-Prigge, S. & Moran, J. V. Genomic deletions created upon LINE-1 retrotransposition. Cell 110, 315–325 (2002)
Moran, J. V. et al. High frequency retrotransposition in cultured mammalian cells. Cell 87, 917–927 (1996)
Ostertag, E. M., Prak, E. T., DeBerardinis, R. J., Moran, J. V. & Kazazian, H. H., Jr Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 28, 1418–1423 (2000)
Goodier, J. L., Ostertag, E. M., Du, K. & Kazazian, H. H., Jr A novel active L1 retrotransposon subfamily in the mouse. Genome Res. 11, 1677–1685 (2001)
Han, J. S. & Boeke, J. D. A highly active synthetic mammalian retrotransposon. Nature 429, 314–318 (2004)
Sugano, T., Kajikawa, M. & Okada, N. Isolation and characterization of retrotransposition-competent LINEs from zebrafish. Gene 365, 74–82 (2006)
Loh, T. P., Sievert, L. L. & Scott, R. W. Proviral sequences that restrict retroviral expression in mouse embryonal carcinoma cells. Mol. Cell. Biol. 7, 3775–3784 (1987)
Teich, N. M., Weiss, R. A., Martin, G. R. & Lowy, D. R. Virus infection of murine teratocarcinoma stem cell lines. Cell 12, 973–982 (1977)
Wolf, D. & Goff, S. P. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 131, 46–57 (2007)
Wolf, D. & Goff, S. P. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458, 1201–1204 (2009)
Li, X. et al. Generation of destabilized green fluorescent protein as a transcription reporter. J. Biol. Chem. 273, 34970–34975 (1998)
Gummow, B. M., Scheys, J. O., Cancelli, V. R. & Hammer, G. D. Reciprocal regulation of a glucocorticoid receptor-steroidogenic factor-1 transcription complex on the Dax-1 promoter by glucocorticoids and adrenocorticotropic hormone in the adrenal cortex. Mol. Endocrinol. 20, 2711–2723 (2006)
Matthaei, K. I., Andrews, P. W. & Bronson, D. L. Retinoic acid fails to induce differentiation in human teratocarcinoma cell lines that express high levels of a cellular receptor protein. Exp. Cell Res. 143, 471–474 (1983)
Kubo, S. et al. L1 retrotransposition in nondividing and primary human somatic cells. Proc. Natl Acad. Sci. USA 103, 8036–8041 (2006)
Bestor, T. H. & Tycko, B. Creation of genomic methylation patterns. Nature Genet. 12, 363–367 (1996)
Bourc'his, D. & Bestor, T. H. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431, 96–99 (2004)
Schumann, G. G. APOBEC3 proteins: major players in intracellular defence against LINE-1-mediated retrotransposition. Biochem. Soc. Trans. 35, 637–642 (2007)
Stetson, D. B., Ko, J. S., Heidmann, T. & Medzhitov, R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134, 587–598 (2008)
Suzuki, J. et al. Genetic evidence that the non-homologous end-joining repair pathway is involved in LINE retrotransposition. PLoS Genet. 5, e1000461 (2009)
Acknowledgements
We thank P. W. Andrews for providing human EC lines, discussing unpublished data from his laboratory and giving advice during the course of this study. We thank A. Macia and M. Munoz-Lopez (Andalusian Stem Cell Bank) for sharing unpublished data; A. V. Furano, H. Kazazian, J. K. Kim, H. Kopera, A. Muotri, and members of the Moran and Garcia-Perez laboratories for critical reading of the manuscript; and G. Smith and L. Villa for help in creating the time-lapsed movie. We thank M. Velkey for providing plasmid pBSSK-pgk; H. Kazazian for providing plasmid pJCC5/LRE3; J. Boeke for providing synthetic mouse LINE-1 constructs; M. Kajikawa and N. Okada for providing the zebrafish LINE-2 expression plasmids; T. Fanning for providing the polyclonal ORF1 antibody; I. Damjanov for comments on the teratoma characterization; T. Lanigan for preparing Moloney murine leukaemia virus retroviral supernatants; C. Pigott for EC culture advice; and T. de la Cueva, P. Catalina and A. Nieto (Andalusian Stem Cell Bank) for their help with mouse experimentation, SKY-FISH and pathology analyses, respectively. J.V.M. is supported by the National Institutes of Health (NIH) (GM060518 and GM082970) and the Howard Hughes Medical Institute. J.L.G.-P. is supported by the Instituto de Salud Carlos III - Consejeria de Salud Junta de Andalucia (ISCIII-CSJA) (EMER07/056), by a Marie Curie International Reintegration Grant action (FP7-PEOPLE-2007-4-3-IRG), by CICE (P09-CTS-4980) and Proyectos en Salud (PI0002/2009) from Junta de Andalucia (Spain) and through the Spanish Ministry of Health (FIS PI08171 and Miguel Servet CP07/00065). M.M. is supported by the ISCIII-CSJA (EMER07/056). P.M. is supported by the Spanish Ministry of Science and Innovation MICINN-PLANE (PLE-2009-0111), by CICE (P08-CTS-3678) from Junta de Andalucia (Spain) and by the Spanish Ministry of Health (FIS PI070026). K.S.O’S. is supported by the NIH (NS-048187 and GM-069985). K.L.C. is supported by the Burroughs Wellcome Foundation and by an NIH Research Project Grant (R01) (AI051198). G.D.H is supported by a National Institute of Diabetes and Digestive and Kidney Diseases NIH R01 (DK62027). J.O.S. is supported by a Cellular and Molecular Approaches to Systems and Integrative Biology Training Grant (T32-GM08322). D.A.K. is supported by The Irvington Institute Fellowship Program of the Cancer Research Institute. S.M. is supported by a CICE (P08-CTS-3678) scholarship from Junta de Andalucia, Spain. C.C.C. is supported by a Rackham Predoctoral Fellowship from the University of Michigan. We defrayed the costs of DNA sequencing in part with the University of Michigan's Cancer Center Support Grant (NIH 5 P30 CA46592).
Author information
Authors and Affiliations
Contributions
J.V.M. and J.L.G.-P. directed the project, designed experiments and drafted the manuscript. J.L.G.-P. performed experiments with the assistance of M.M. and K.S.O’S. (cell cycle experiments), J.O.S. and G.D.H (chromatin immunoprecipitation experiments), D.A.K., C.C.C. and K.L.C. (human immunodeficiency virus-based experiments), and S.M. and P.M. (teratoma assays). All the authors commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Methods, legend for Supplementary Movie 1, Supplementary Tables 1-2, Supplementary Figures 1-10 with legends and References. (PDF 17434 kb)
Supplementary Movie 1
This movie shows reactivation of L1-retro-EGFP expression in pk-5 Cells (see page 17 of the Supplementary Information file for full legend). (MOV 962 kb)
Rights and permissions
About this article
Cite this article
Garcia-Perez, J., Morell, M., Scheys, J. et al. Epigenetic silencing of engineered L1 retrotransposition events in human embryonic carcinoma cells. Nature 466, 769–773 (2010). https://doi.org/10.1038/nature09209
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature09209
This article is cited by
-
Fanconi anemia DNA crosslink repair factors protect against LINE-1 retrotransposition during mouse development
Nature Structural & Molecular Biology (2023)
-
Retrotransposon activation during Drosophila metamorphosis conditions adult antiviral responses
Nature Genetics (2022)
-
Unbiased proteomic mapping of the LINE-1 promoter using CRISPR Cas9
Mobile DNA (2021)
-
Characterization of an active LINE-1 in the naked mole-rat genome
Scientific Reports (2021)
-
Stabilization of heterochromatin by CLOCK promotes stem cell rejuvenation and cartilage regeneration
Cell Research (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.