Abstract
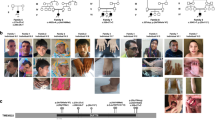
The development of the human cerebral cortex is an orchestrated process involving the generation of neural progenitors in the periventricular germinal zones, cell proliferation characterized by symmetric and asymmetric mitoses, followed by migration of post-mitotic neurons to their final destinations in six highly ordered, functionally specialized layers1,2. An understanding of the molecular mechanisms guiding these intricate processes is in its infancy, substantially driven by the discovery of rare mutations that cause malformations of cortical development3,4,5,6. Mapping of disease loci in putative Mendelian forms of malformations of cortical development has been hindered by marked locus heterogeneity, small kindred sizes and diagnostic classifications that may not reflect molecular pathogenesis. Here we demonstrate the use of whole-exome sequencing to overcome these obstacles by identifying recessive mutations in WD repeat domain 62 (WDR62) as the cause of a wide spectrum of severe cerebral cortical malformations including microcephaly, pachygyria with cortical thickening as well as hypoplasia of the corpus callosum. Some patients with mutations in WDR62 had evidence of additional abnormalities including lissencephaly, schizencephaly, polymicrogyria and, in one instance, cerebellar hypoplasia, all traits traditionally regarded as distinct entities. In mice and humans, WDR62 transcripts and protein are enriched in neural progenitors within the ventricular and subventricular zones. Expression of WDR62 in the neocortex is transient, spanning the period of embryonic neurogenesis. Unlike other known microcephaly genes, WDR62 does not apparently associate with centrosomes and is predominantly nuclear in localization. These findings unify previously disparate aspects of cerebral cortical development and highlight the use of whole-exome sequencing to identify disease loci in settings in which traditional methods have proved challenging.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Bystron, I., Blakemore, C. & Rakic, P. Development of the human cerebral cortex: Boulder Committee revisited. Nature Rev. Neurosci. 9, 110–122 (2008)
Rakic, P. Evolution of the neocortex: a perspective from developmental biology. Nature Rev. Neurosci. 10, 724–735 (2009)
Guerrini, R., Dobyns, W. B. & Barkovich, A. J. Abnormal development of the human cerebral cortex: genetics, functional consequences and treatment options. Trends Neurosci. 31, 154–162 (2008)
Guerrini, R. Genetic malformations of the cerebral cortex and epilepsy. Epilepsia 46 (suppl. 1). 32–37 (2005)
Guerrini, R. & Carrozzo, R. Epilepsy and genetic malformations of the cerebral cortex. Am. J. Med. Genet. 106, 160–173 (2001)
Mochida, G. H. & Walsh, C. A. Molecular genetics of human microcephaly. Curr. Opin. Neurol. 14, 151–156 (2001)
Barkovich, A. J., Kuzniecky, R. I., Jackson, G. D., Guerrini, R. & Dobyns, W. B. Classification system for malformations of cortical development: update 2001. Neurology 57, 2168–2178 (2001)
Barkovich, A. J., Kuzniecky, R. I., Jackson, G. D., Guerrini, R. & Dobyns, W. B. A developmental and genetic classification for malformations of cortical development. Neurology 65, 1873–1887 (2005)
Choi, M. et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc. Natl Acad. Sci. USA 106, 19096–19101 (2009)
Ng, S. B. et al. Exome sequencing identifies the cause of a mendelian disorder. Nature Genet. 42, 30–35 (2010)
Ng, S. B. et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature 461, 272–276 (2009)
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007)
Wasserman, T. et al. A novel c-Jun N-terminal kinase (JNK)-binding protein WDR62 is recruited to stress granules and mediates a nonclassical JNK activation. Mol. Biol. Cell 21, 117–130 (2010)
Bond, J. et al. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nature Genet. 37, 353–355 (2005)
Kumar, A., Girimaji, S. C., Duvvari, M. R. & Blanton, S. H. Mutations in STIL, encoding a pericentriolar and centrosomal protein, cause primary microcephaly. Am. J. Hum. Genet. 84, 286–290 (2009)
Thornton, G. K. & Woods, C. G. Primary microcephaly: do all roads lead to Rome? Trends Genet. 25, 501–510 (2009)
Roberts, E. et al. The second locus for autosomal recessive primary microcephaly (MCPH2) maps to chromosome 19q13.1–13.2. Eur. J. Hum. Genet. 7, 815–820 (1999)
Hartl, D. et al. Transcriptome and proteome analysis of early embryonic mouse brain development. Proteomics 8, 1257–1265 (2008)
Li, H., Ruan, J. & Durbin, R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18, 1851–1858 (2008)
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009)
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009)
Irizarry, R. A. et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31, e15 (2003)
Huang da, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 4, 44–57 (2009)
Stillman, A. A. et al. Developmentally regulated and evolutionarily conserved expression of SLITRK1 in brain circuits implicated in Tourette syndrome. J. Comp. Neurol. 513, 21–37 (2009)
Abelson, J. F. et al. Sequence variants in SLITRK1 are associated with Tourette’s syndrome. Science 310, 317–320 (2005)
Kwan, K. Y. et al. SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc. Natl Acad. Sci. USA 105, 16021–16026 (2008)
Acknowledgements
We are indebted to the patients and families who have contributed to this study. We thank J. Noonan for expert advice and C. Camputaro for her help with three-dimensional reconstruction of the magnetic resonance images. This study was supported by the Yale Program on Neurogenetics, the Yale Center for Human Genetics and Genomics, and National Institutes of Health grants RC2 NS070477 (to M.G.), UL1 RR024139NIH (Yale Clinical and Translational Science Award) and UO1MH081896 (to N.S.). SNP genotyping was supported in part by a National Institutes of Health Neuroscience Microarray Consortium award U24 NS051869-02S1 (to S.M.). R.P.L. is an investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
M.W.S., R.P.L. and M.G. designed the study and K.B., A.L., N.S., R.P.L. and M.G. designed the experiments. K.B., A.K.O., A.L., K.Y.K., T.B., M.B., S.S., W.H. and S.M. performed the experiments. B.T., D.Y., B.T., A.O.C., S.G., H.K., S.Y., H.P., C.Y., S.K., M.T. and M.O. identified, consented and recruited the study subjects and provided clinical information. A.D., M.H.J., R.A.B., N.K. and M.N.P. performed and evaluated magnetic resonance imaging. M.C. and R.P.L. developed the bioinformatics scripts for data analysis. K.B., A.K.O., K.Y., A.L. and M.G. analysed the genetics data. A.L., K.Y.K, Y.Z., N.S. and M.G. analysed the expression data. K.B., A.K.O, A.L., R.P.L., M.W.S. and M.G. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors have a provisional patent application under consideration based on the findings of this work. R.A.B. is a consultant for Bristol-Myers Squibb.
Supplementary information
Supplementary Information
This file contains Supplementary Patient Notes, legends for Supplementary Videos 1- 6 (see separate Movie files 1-6), Supplementary Figures 1-8 with legends and Supplementary Tables 1-6. (PDF 2092 kb)
Supplementary Movie 1
This video is constructed from T2 sagittal images (photographically inverted) of patient NG 26-1 and demonstrates microcephaly and cortical thickening (see Supplementary Information file for full legend). (MOV 4003 kb)
Supplementary Movie 2
This video is constructed from T2 coronal images (photographically inverted) of patient NG 26-1 and demonstrates microcephaly and cortical thickening (see Supplementary Information file for full legend). (MOV 3620 kb)
Supplementary Movie 3
This video is constructed from T2 sagittal images of patient NG 190-1 (photographically inverted) and it shows diffuse cortical volume loss and marked craniofacial disproportion (see Supplementary Information file for full legend). (MOV 3957 kb)
Supplementary Movie 4
This video is constructed from T2 coronal images of patient NG 190-1 (photographically inverted) and it shows diffuse cortical volume loss and marked craniofacial disproportion (see Supplementary Information file for full legend). (MOV 3672 kb)
Supplementary Movie 5
This video is constructed from T1 sagittal images (photographically inverted) of patient NG 891-1 and it demonstrates radiographic findings consistent with microlissencephaly including prominent microcephaly (see Supplementary Information file for full legend). (MOV 716 kb)
Supplementary Movie 6
This video is constructed from T1 coronal images (photographically inverted) of patient NG 891-1 and it demonstrates radiographic findings consistent with microlissencephaly including prominent microcephaly (see Supplementary Information file for full legend). (MOV 119 kb)
Rights and permissions
About this article
Cite this article
Bilgüvar, K., Öztürk, A., Louvi, A. et al. Whole-exome sequencing identifies recessive WDR62 mutations in severe brain malformations. Nature 467, 207–210 (2010). https://doi.org/10.1038/nature09327
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09327
This article is cited by
-
WDR62-deficiency Causes Autism-like Behaviors Independent of Microcephaly in Mice
Neuroscience Bulletin (2023)
-
Mutation of key signaling regulators of cerebrovascular development in vein of Galen malformations
Nature Communications (2023)
-
Ophthalmic genetic counselling: emerging trends in practice perspectives in Asia
Journal of Community Genetics (2022)
-
Novel phenotype and genotype spectrum of WDR62 in two patients with associated primary autosomal recessive microcephaly
Irish Journal of Medical Science (1971 -) (2022)
-
A case report of microcephaly and refractory West syndrome associated with WDR62 mutation
Acta Epileptologica (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.