Abstract
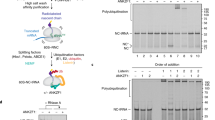
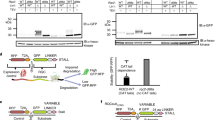
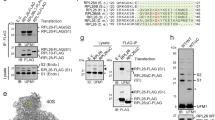
Messenger RNA lacking stop codons (‘non-stop mRNA’) can arise from errors in gene expression, and encode aberrant proteins whose accumulation could be deleterious to cellular function1,2. In bacteria, these ‘non-stop proteins’ become co-translationally tagged with a peptide encoded by ssrA/tmRNA (transfer-messenger RNA), which signals their degradation by energy-dependent proteases1,3. How eukaryotic cells eliminate non-stop proteins has remained unknown. Here we show that the Saccharomyces cerevisiae Ltn1 RING-domain-type E3 ubiquitin ligase acts in the quality control of non-stop proteins, in a process that is mechanistically distinct but conceptually analogous to that performed by ssrA: Ltn1 is predominantly associated with ribosomes, and it marks nascent non-stop proteins with ubiquitin to signal their proteasomal degradation. Ltn1-mediated ubiquitylation of non-stop proteins seems to be triggered by their stalling in ribosomes on translation through the poly(A) tail. The biological relevance of this process is underscored by the finding that loss of Ltn1 function confers sensitivity to stress caused by increased non-stop protein production. We speculate that defective protein quality control may underlie the neurodegenerative phenotype that results from mutation of the mouse Ltn1 homologue Listerin.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Moore, S. D. & Sauer, R. T. The tmRNA system for translational surveillance and ribosome rescue. Annu. Rev. Biochem. 76, 101–124 (2007)
Frischmeyer, P. A. et al. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295, 2258–2261 (2002)
Dulebohn, D. et al. Trans-translation: the tmRNA-mediated surveillance mechanism for ribosome rescue, directed protein degradation, and nonstop mRNA decay. Biochemistry 46, 4681–4693 (2007)
van Hoof, A. et al. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295, 2262–2264 (2002)
Doma, M. K. & Parker, R. RNA quality control in eukaryotes. Cell 131, 660–668 (2007)
Wilson, M. A., Meaux, S. & van Hoof, A. A genomic screen in yeast reveals novel aspects of nonstop mRNA metabolism. Genetics 177, 773–784 (2007)
Ito-Harashima, S. et al. Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev. 21, 519–524 (2007)
Li, W. et al. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS ONE 3, e1487 (2008)
Deshaies, R. J. & Joazeiro, C. A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 (2009)
Chu, J. et al. A mouse forward genetics screen identifies LISTERIN as an E3 ubiquitin ligase involved in neurodegeneration. Proc. Natl Acad. Sci. USA 106, 2097–2103 (2009)
Braun, M. A. et al. Identification of Rkr1, a nuclear RING domain protein with functional connections to chromatin modification in Saccharomyces cerevisiae . Mol. Cell. Biol. 27, 2800–2811 (2007)
Inada, T. & Aiba, H. Translation of aberrant mRNAs lacking a termination codon or with a shortened 3′-UTR is repressed after initiation in yeast. EMBO J. 24, 1584–1595 (2005)
Verma, R. et al. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell 11, 3425–3439 (2000)
McClellan, A. J., Scott, M. D. & Frydman, J. Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell 121, 739–748 (2005)
Dimitrova, L. N. et al. Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J. Biol. Chem. 284, 10343–10352 (2009)
Lu, J. & Deutsch, C. Electrostatics in the ribosomal tunnel modulate chain elongation rates. J. Mol. Biol. 384, 73–86 (2008)
Fleischer, T. C. et al. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev. 20, 1294–1307 (2006)
Brodersen, D. E. et al. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 103, 1143–1154 (2000)
Kirillov, S. et al. Movement of the 3′-end of tRNA through the peptidyl transferase centre and its inhibition by antibiotics. FEBS Lett. 406, 223–233 (1997)
Bradley, M. E. et al. Guanidine reduces stop codon read-through caused by missense mutations in SUP35 or SUP45. Yeast 20, 625–632 (2003)
Rospert, S. Ribosome function: governing the fate of a nascent polypeptide. Curr. Biol. 14, R386–R388 (2004)
Ghaemmaghami, S. et al. Global analysis of protein expression in yeast. Nature 425, 737–741 (2003)
Zenklusen, D., Larson, D. R. & Singer, R. H. Single-RNA counting reveals alternative modes of gene expression in yeast. Nature Struct. Mol. Biol. 15, 1263–1271 (2008)
Acknowledgements
We thank M. C. Sogayar for continued mentorship; I. Kwan for technical help; A. Bacconi for help with microscopy; T. Inada, A. van Hoof, S. Liebman and J. Frydman for reagents; the Williamson and Saez laboratories for help with sucrose-gradient fractionation and real-time PCR, respectively; E. Peters for mass spectrometry analyses; M. Smolka for help with yeast methods; and R. Deshaies, E. P. Geiduschek and T. Hunter for discussions. Work in the Joazeiro laboratory is supported by Research Scholar Grant 08-298-01-TBE from the American Cancer Society and grant R01GM083060 from the National Institute of General Medical Sciences.
Author information
Authors and Affiliations
Contributions
C.A.P.J. and M.H.B. designed the studies, interpreted the data and wrote the manuscript. M.H.B. conducted the experiments.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Methods, a List of Genotypes, Supplementary Figures 1-9 with legends, Supplementary Table 1, and additional references. (PDF 3181 kb)
Rights and permissions
About this article
Cite this article
Bengtson, M., Joazeiro, C. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature 467, 470–473 (2010). https://doi.org/10.1038/nature09371
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09371
This article is cited by
-
Crosstalk between protein post-translational modifications and phase separation
Cell Communication and Signaling (2024)
-
Decoding of the ubiquitin code for clearance of colliding ribosomes by the RQT complex
Nature Communications (2023)
-
Remodeling of the ribosomal quality control and integrated stress response by viral ubiquitin deconjugases
Nature Communications (2023)
-
Molecular basis for recognition and deubiquitination of 40S ribosomes by Otu2
Nature Communications (2023)
-
Presence of immunogenic alternatively spliced insulin gene product in human pancreatic delta cells
Diabetologia (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.