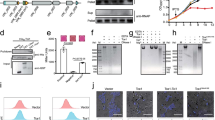
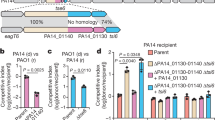
Abstract
Bacteria have developed mechanisms to communicate and compete with one another in diverse environments1. A new form of intercellular communication, contact-dependent growth inhibition (CDI), was discovered recently in Escherichia coli2. CDI is mediated by the CdiB/CdiA two-partner secretion (TPS) system. CdiB facilitates secretion of the CdiA ‘exoprotein’ onto the cell surface. An additional small immunity protein (CdiI) protects CDI+ cells from autoinhibition2,3. The mechanisms by which CDI blocks cell growth and by which CdiI counteracts this growth arrest are unknown. Moreover, the existence of CDI activity in other bacteria has not been explored. Here we show that the CDI growth inhibitory activity resides within the carboxy-terminal region of CdiA (CdiA-CT), and that CdiI binds and inactivates cognate CdiA-CT, but not heterologous CdiA-CT. Bioinformatic and experimental analyses show that multiple bacterial species encode functional CDI systems with high sequence variability in the CdiA-CT and CdiI coding regions. CdiA-CT heterogeneity implies that a range of toxic activities are used during CDI. Indeed, CdiA-CTs from uropathogenic E. coli and the plant pathogen Dickeya dadantii have different nuclease activities, each providing a distinct mechanism of growth inhibition. Finally, we show that bacteria lacking the CdiA-CT and CdiI coding regions are unable to compete with isogenic wild-type CDI+ cells both in laboratory media and on a eukaryotic host. Taken together, these results suggest that CDI systems constitute an intricate immunity network with an important function in bacterial competition.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hibbing, M. E., Fuqua, C., Parsek, M. R. & Peterson, S. B. Bacterial competition: surviving and thriving in the microbial jungle. Nature Rev. Microbiol. 8, 15–25 (2010)
Aoki, S. K. et al. Contact-dependent inhibition of growth in Escherichia coli. Science 309, 1245–1248 (2005)
Aoki, S. K. et al. Contact-dependent growth inhibition requires the essential outer membrane protein BamA (YaeT) as the receptor and the inner membrane transport protein AcrB. Mol. Microbiol. 70, 323–340 (2008)
Aoki, S. K., Webb, J. S., Braaten, B. A. & Low, D. A. Contact-dependent growth inhibition causes reversible metabolic downregulation in Escherichia coli. J. Bacteriol. 191, 1777–1786 (2009)
Luo, C., Hu, G. Q. & Zhu, H. Genome reannotation of Escherichia coli CFT073 with new insights into virulence. BMC Genomics 10, 552–561 (2009)
Pagni, M. & Jongeneel, C. V. Making sense of score statistics for sequence alignments. Brief. Bioinform. 2, 51–67 (2001)
Dobrindt, U. et al. Genetic structure and distribution of four pathogenicity islands (PAI I536 to PAI IV536) of uropathogenic Escherichia coli strain 536. Infect. Immun. 70, 6365–6372 (2002)
Karimova, G., Pidoux, J., Ullmann, A. & Ladant, D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl Acad. Sci. USA 95, 5752–5756 (1998)
Duport, C., Baysse, C. & Michel-Briand, Y. Molecular characterization of pyocin S3, a novel S-type pyocin from Pseudomonas aeruginosa. J. Biol. Chem. 270, 8920–8927 (1995)
Rojas, C. M. et al. The Erwinia chrysanthemi EC16 hrp/hrc gene cluster encodes an active Hrp type III secretion system that is flanked by virulence genes functionally unrelated to the Hrp system. Mol. Plant Microbe Interact. 17, 644–653 (2004)
Ham, J. H. et al. Analysis of Erwinia chrysanthemi EC16 pelE:uidA, pelL:uidA, and hrpN:uidA mutants reveals strain-specific atypical regulation of the Hrp type III secretion system. Mol. Plant Microbe Interact. 17, 184–194 (2004)
Yap, M. N., Rojas, C. M., Yang, C. H. & Charkowski, A. O. Harpin mediates cell aggregation in Erwinia chrysanthemi 3937. J. Bacteriol. 188, 2280–2284 (2006)
Edwards, R. A., Keller, L. H. & Schifferli, D. M. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207, 149–157 (1998)
Braaten, B. A., Nou, X., Kaltenbach, L. S. & Low, D. A. Methylation patterns in pap regulatory DNA control pyelonephritis-associated pili phase variation in E.coli. Cell 76, 577–588 (1994)
Acknowledgements
We thank A. Charkowski, A. Collmer, J. Roth and H. Schweizer for plasmids, bacterial strains and helpful discussions; W. Lathem for Y. pestis CO92 DNA; and R. Christoffersen for helpful advice on plant experiments. This work was supported by National Science Foundation grant 0642052 (D.A.L.), a Tri-Counties Blood Bank Postdoctoral Fellowship (S.K.A.) and National Institutes of Health grants GM078634 (C.S.H.), AI043986 (P.A.C.) and U54AI065359 (D.A.L., P.A.C. and C.S.H.). The content is the sole responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health. This project made use of preliminary sequences from the Dickeya dadantii 3937 genome project supported by the Initiative for Future Agriculture and Food Systems grant no. 2001-52100-11316 from the United States Department of Agriculture Cooperative State Research, Education, and Extension Service.
Author information
Authors and Affiliations
Contributions
D.A.L., S.K.A., P.A.C. and C.S.H. designed the research. D.A.L., C.S.H., P.A.C., S.J.P. and S.K.A. prepared the manuscript. S.J.P. and B.R.B. performed bioinformatic analyses. B.R.B., A.M.J. and P.A.C. obtained initial evidence for the toxic nature of CdiA-CT, variability of CdiA-CTs and CdiIs, and binding between CdiA-CTs and cognate CdiIs in studies of Burkholderia pseudomallei cdi genes. S.K.A. cloned and performed competition assays and growth curves involving cdiBAI, cdiA-CT and cdiA chimaeras and deletions. B.A.B. and J.S.W. conducted the deletion mapping study, with assistance from S.K.A. B.A.B. and S.K.A. conducted the bacterial two-hybrid study. C.T.R. constructed D. dadantii cdi mutants and plasmids and performed growth competition assays on chicory. E.J.D. cloned, purified protein and performed the in vitro protein interaction and CdiA-CT activity studies.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-4 with legends, Supplementary Methods, Supplementary Tables 1-2 and additional references. (PDF 1393 kb)
Rights and permissions
About this article
Cite this article
Aoki, S., Diner, E., de Roodenbeke, C. et al. A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature 468, 439–442 (2010). https://doi.org/10.1038/nature09490
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09490
This article is cited by
-
Structural basis for the toxic activity of MafB2 from maf genomic island 2 (MGI-2) in N. meningitidis B16B6
Scientific Reports (2023)
-
Bacterial defences: mechanisms, evolution and antimicrobial resistance
Nature Reviews Microbiology (2023)
-
The control of waterborne pathogenic bacteria in fresh water using a biologically active filter
npj Clean Water (2022)
-
Proteolytic processing induces a conformational switch required for antibacterial toxin delivery
Nature Communications (2022)
-
The extracellular contractile injection system is enriched in environmental microbes and associates with numerous toxins
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.