Abstract
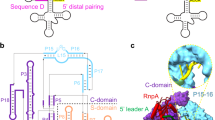
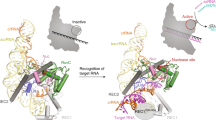
Ribonuclease (RNase) P is the universal ribozyme responsible for 5′-end tRNA processing. We report the crystal structure of the Thermotoga maritima RNase P holoenzyme in complex with tRNAPhe. The 154 kDa complex consists of a large catalytic RNA (P RNA), a small protein cofactor and a mature tRNA. The structure shows that RNA–RNA recognition occurs through shape complementarity, specific intermolecular contacts and base-pairing interactions. Soaks with a pre-tRNA 5′ leader sequence with and without metal help to identify the 5′ substrate path and potential catalytic metal ions. The protein binds on top of a universally conserved structural module in P RNA and interacts with the leader, but not with the mature tRNA. The active site is composed of phosphate backbone moieties, a universally conserved uridine nucleobase, and at least two catalytically important metal ions. The active site structure and conserved RNase P–tRNA contacts suggest a universal mechanism of catalysis by RNase P.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Hartmann, R. K., Gossringer, M., Spath, B., Fischer, S. & Marchfelder, A. The making of tRNAs and more – RNase P and tRNase Z. Prog. Mol. Biol. Transl. Sci. 85, 319–368 (2009)
Kazantsev, A. V. & Pace, N. R. Bacterial RNase P: a new view of an ancient enzyme. Nature Rev. Microbiol. 4, 729–740 (2006)
Liu, F. & Altman, S. Protein Reviews Volume 10: Ribonuclease P (Springer, 2010)
Guerrier-Takada, C., Gardiner, K., Marsh, T., Pace, N. & Altman, S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 35, 849–857 (1983)
Pan, T., Loria, A. & Zhong, K. Probing of tertiary interactions in RNA: 2′-hydroxyl-base contacts between the RNase P RNA and pre-tRNA. Proc. Natl Acad. Sci. USA 92, 12510–12514 (1995)
Hsieh, J. et al. A divalent cation stabilizes the active conformation of the B. subtilis RNase P pre-tRNA complex: a role for an inner-sphere metal ion in RNase P. J. Mol. Biol. 400, 38–51 (2010)
Kirsebom, L. A. in Protein Reviews Volume 10: RNase P (eds Liu, F. & Altman, S. ) Ch. 7, 113–134 (Springer, 2010)
Buck, A. H., Dalby, A. B., Poole, A. W., Kazantsev, A. V. & Pace, N. R. Protein activation of a ribozyme: the role of bacterial RNase P protein. EMBO J. 24, 3360–3368 (2005)
Kurz, J. C., Niranjanakumari, S. & Fierke, C. A. Protein component of Bacillus subtilis RNase P specifically enhances the affinity for precursor-tRNAAsp . Biochemistry 37, 2393–2400 (1998)
Peck-Miller, K. A. & Altman, S. Kinetics of the processing of the precursor to 4.5 S RNA, a naturally occurring substrate for RNase P from Escherichia coli . J. Mol. Biol. 221, 1–5 (1991)
Koutmou, K. S. et al. Protein-precursor tRNA contact leads to sequence-specific recognition of 5′ leaders by bacterial ribonuclease P. J. Mol. Biol. 396, 195–208 (2010)
Sun, L., Campbell, F. E., Zahler, N. H. & Harris, M. E. Evidence that substrate-specific effects of C5 protein lead to uniformity in binding and catalysis by RNase P. EMBO J. 25, 3998–4007 (2006)
Reich, C., Olsen, G. J., Pace, B. & Pace, N. R. Role of the protein moiety of ribonuclease P, a ribonucleoprotein enzyme. Science 239, 178–181 (1988)
Kazantsev, A. V. et al. Crystal structure of a bacterial ribonuclease P RNA. Proc. Natl Acad. Sci. USA 102, 13392–13397 (2005)
Krasilnikov, A. S., Xiao, Y., Pan, T. & Mondragón, A. Basis for structural diversity in homologous RNAs. Science 306, 104–107 (2004)
Krasilnikov, A. S., Yang, X., Pan, T. & Mondragón, A. Crystal structure of the specificity domain of ribonuclease P. Nature 421, 760–764 (2003)
Torres-Larios, A., Swinger, K. K., Krasilnikov, A. S., Pan, T. & Mondragón, A. Crystal structure of the RNA component of bacterial ribonuclease P. Nature 437, 584–587 (2005)
Chen, J.-L. & Pace, N. R. Identification of the universally conserved core of ribonuclease P RNA. RNA 3, 557–560 (1997)
Torres-Larios, A., Swinger, K. K., Pan, T. & Mondragón, A. Structure of ribonuclease P–a universal ribozyme. Curr. Opin. Struct. Biol. 16, 327–335 (2006)
Ferré-d’Amaré, A. R., Zhou, K. & Doudna, J. A. A general module for RNA crystallization. J. Mol. Biol. 279, 621–631 (1998)
Kazantsev, A. V. et al. High-resolution structure of RNase P protein from Thermotoga maritima . Proc. Natl Acad. Sci. USA 100, 7497–7502 (2003)
Shi, H. & Moore, P. B. The crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution: a classic structure revisited. RNA 6, 1091–1105 (2000)
Kirsebom, L. A. & Svard, S. G. Base pairing between Escherichia coli RNase P RNA and its substrate. EMBO J. 13, 4870–4876 (1994)
Stams, T., Niranjanakumari, S., Fierke, C. A. & Christianson, D. W. Ribonuclease P protein structure: evolutionary origins in the translational apparatus. Science 280, 752–755 (1998)
Massire, C., Jaeger, L. & Westhof, E. Derivation of the three-dimensional architecture of bacterial ribonuclease P RNAs from comparative sequence analysis. J. Mol. Biol. 279, 773–793 (1998)
Buck, A. H., Kazantsev, A. V., Dalby, A. B. & Pace, N. R. Structural perspective on the activation of RNase P RNA by protein. Nature Struct. Mol. Biol. 12, 958–964 (2005)
Tsai, H. Y., Masquida, B., Biswas, R., Westhof, E. & Gopalan, V. Molecular modeling of the three-dimensional structure of the bacterial RNase P holoenzyme. J. Mol. Biol. 325, 661–675 (2003)
LaGrandeur, T. E., Huttenhofer, A., Noller, H. F. & Pace, N. R. Phylogenetic comparative chemical footprint analysis of the interaction between ribonuclease P RNA and tRNA. EMBO J. 13, 3945–3952 (1994)
Mondragón, A. in Protein Reviews Volume 10: Ribonuclease P (eds Liu, F. & Altman, S. ) Ch. 4, 63–78 (Springer, 2010)
Biswas, R., Ledman, D. W., Fox, R. O., Altman, S. & Gopalan, V. Mapping RNA-protein interactions in ribonuclease P from Escherichia coli using disulfide-linked EDTA-Fe. J. Mol. Biol. 296, 19–31 (2000)
Niranjanakumari, S., Stams, T., Crary, S. M., Christianson, D. W. & Fierke, C. A. Protein component of the ribozyme ribonuclease P alters substrate recognition by directly contacting precursor tRNA. Proc. Natl Acad. Sci. USA 95, 15212–15217 (1998)
Christian, E. L., Kaye, N. M. & Harris, M. E. Helix P4 is a divalent metal ion binding site in the conserved core of the ribonuclease P ribozyme. RNA 6, 511–519 (2000)
Crary, S. M., Kurz, J. C. & Fierke, C. A. Specific phosphorothioate substitutions probe the active site of Bacillus subtilis ribonuclease P. RNA 8, 933–947 (2002)
Christian, E. L., Smith, K. M., Perera, N. & Harris, M. E. The P4 metal binding site in RNase P RNA affects active site metal affinity through substrate positioning. RNA 12, 1463–1467 (2006)
Kazantsev, A. V., Krivenko, A. A. & Pace, N. R. Mapping metal-binding sites in the catalytic domain of bacterial RNase P RNA. RNA 15, 266–276 (2009)
Pomeranz Krummel, D. A., Kent, O., MacMillan, A. M. & Altman, S. Evidence for helical unwinding of an RNA substrate by the RNA enzyme RNase P: use of an interstrand disulfide crosslink in substrate. J. Mol. Biol. 295, 1113–1118 (2000)
Gaur, R. K., Hanne, A., Conrad, F., Kahle, D. & Krupp, G. Differences in the interaction of Escherichia coli RNase P RNA with tRNAs containing a short or a long extra arm. RNA 2, 674–681 (1996)
Forster, A. C. & Altman, S. Similar cage-shaped structures for the RNA components of all ribonuclease P and ribonuclease MRP enzymes. Cell 62, 407–409 (1990)
Burkard, U. & Soll, D. The unusually long amino acid acceptor stem of Escherichia coli selenocysteine tRNA results from abnormal cleavage by RNase P. Nucleic Acids Res. 16, 11617–11624 (1988)
Walker, S. C. & Engelke, D. R. Ribonuclease P: the evolution of an ancient RNA enzyme. Crit. Rev. Biochem. Mol. Biol. 41, 77–102 (2006)
Stahley, M. R. & Strobel, S. A. Structural evidence for a two-metal-ion mechanism of group I intron splicing. Science 309, 1587–1590 (2005)
Toor, N., Keating, K. S., Taylor, S. D. & Pyle, A. M. Crystal structure of a self-spliced group II intron. Science 320, 77–82 (2008)
Steitz, T. A. & Steitz, J. A. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl Acad. Sci. USA 90, 6498–6502 (1993)
Krivenko, A. A., Kazantsev, A. V., Adamidi, C., Harrington, D. J. & Pace, N. R. Expression, purification, crystallization and preliminary diffraction analysis of RNase P protein from Thermotoga maritima . Acta Crystallogr. D 58, 1234–1236 (2002)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007)
de La Fortelle, E. & Bricogne, G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 276, 472–494 (1997)
DeLano, W. L. The PyMOL molecular graphics system. 〈http://www.pymol.org〉 (2002)
Landau, M. et al. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 33, W299–W302 (2005)
Chen, Y., Li, X. & Gegenheimer, P. Ribonuclease P catalysis requires Mg2+ coordinated to the pro-R P oxygen of the scissile bond. Biochemistry 36, 2425–2438 (1997)
Warnecke, J. M. et al. Ribonuclease P (RNase P) RNA is converted to a Cd2+-ribozyme by a single Rp-phosphorothioate modification in the precursor tRNA at the RNase P cleavage site. Proc. Natl Acad. Sci. USA 93, 8924–8928 (1996)
Milligan, J. F., Groebe, D. R., Witherell, G. W. & Uhlenbeck, O. C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 15, 8783–8798 (1987)
Leslie, A. G. W. in Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. 26, 27–33 (1992)
Kabsch, W. Automatic indexing of rotation diffraction patterns. J. Appl. Cryst. 21, 67–72 (1988)
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Strong, M. et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis . Proc. Natl Acad. Sci. USA 103, 8060–8065 (2006)
Abrahams, J. P. & Leslie, A. G. W. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D 52, 30–42 (1996)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Blanc, E. et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D 60, 2210–2221 (2004)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Kleywegt, G. J. Use of non-crystallographic symmetry in protein structure refinement. Acta Crystallogr. D 52, 842–857 (1996)
Acknowledgements
We thank E. Sontheimer and O. Uhlenbeck for comments and suggestions, N. Pace for the gift of the T. maritima RNase P protein plasmid, Obiter Research, A. Davis and M. E. Duban for advice and preparation of iridium hexammine, and A. Samelson for discussions and assistance. In addition, we are grateful for data collection assistance from S. Anderson, Z. Wawrzak, and staff at LS-CAT. Research was supported by the NIH. N.J.R. is an NRSA postdoctoral fellow.
Author information
Authors and Affiliations
Contributions
A.M. directed the work. A.T.-L. and A.M. conceived the project. All authors performed and designed experiments. N.J.R. obtained crystallographic data. N.J.R. and A.M. analysed the crystallographic data. N.J.R. and A.M. wrote the paper with contributions from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-5, Supplementary Figures 1-17 with legends and additional references. (PDF 5600 kb)
Rights and permissions
About this article
Cite this article
Reiter, N., Osterman, A., Torres-Larios, A. et al. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature 468, 784–789 (2010). https://doi.org/10.1038/nature09516
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09516
This article is cited by
-
Importance of residue 248 in Escherichia coli RNase P RNA mediated cleavage
Scientific Reports (2023)
-
Crystal structures and insights into precursor tRNA 5’-end processing by prokaryotic minimal protein-only RNase P
Nature Communications (2022)
-
Cryo-EM structure of catalytic ribonucleoprotein complex RNase MRP
Nature Communications (2020)
-
High-affinity recognition of specific tRNAs by an mRNA anticodon-binding groove
Nature Structural & Molecular Biology (2019)
-
Trapping of the transport-segment DNA by the ATPase domains of a type II topoisomerase
Nature Communications (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.