Abstract
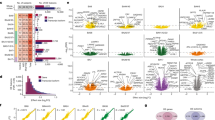
Autism spectrum disorder (ASD) is a common, highly heritable neurodevelopmental condition characterized by marked genetic heterogeneity1,2,3. Thus, a fundamental question is whether autism represents an aetiologically heterogeneous disorder in which the myriad genetic or environmental risk factors perturb common underlying molecular pathways in the brain4. Here, we demonstrate consistent differences in transcriptome organization between autistic and normal brain by gene co-expression network analysis. Remarkably, regional patterns of gene expression that typically distinguish frontal and temporal cortex are significantly attenuated in the ASD brain, suggesting abnormalities in cortical patterning. We further identify discrete modules of co-expressed genes associated with autism: a neuronal module enriched for known autism susceptibility genes, including the neuronal specific splicing factor A2BP1 (also known as FOX1), and a module enriched for immune genes and glial markers. Using high-throughput RNA sequencing we demonstrate dysregulated splicing of A2BP1-dependent alternative exons in the ASD brain. Moreover, using a published autism genome-wide association study (GWAS) data set, we show that the neuronal module is enriched for genetically associated variants, providing independent support for the causal involvement of these genes in autism. In contrast, the immune-glial module showed no enrichment for autism GWAS signals, indicating a non-genetic aetiology for this process. Collectively, our results provide strong evidence for convergent molecular abnormalities in ASD, and implicate transcriptional and splicing dysregulation as underlying mechanisms of neuronal dysfunction in this disorder.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Durand, C. M. et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nature Genet. 39, 25–27 (2006)
Pinto, D. et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature 466, 368–372 (2010)
Sebat, J. et al. Strong association of de novo copy number mutations with autism. Science 316, 445–449 (2007)
Geschwind, D. H. Autism: many genes, common pathways? Cell 135, 391–395 (2008)
Amaral, D. G., Schumann, C. M. & Nordahl, C. W. Neuroanatomy of autism. Trends Neurosci. 31, 137–145 (2008)
Abrahams, B. S. & Geschwind, D. H. Advances in autism genetics: on the threshold of a new neurobiology. Nature Rev. Genet. 9, 341–355 (2008)
Garbett, K. et al. Immune transcriptome alterations in the temporal cortex of subjects with autism. Neurobiol. Dis. 30, 303–311 (2008)
Oldham, M. C. et al. Functional organization of the transcriptome in human brain. Nature Neurosci. 11, 1271–1282 (2008)
Zhang, B. & Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 4, 17 (2005).
Johnson, M. B. et al. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron 62, 494–509 (2009)
Zikopoulos, B. & Barbas, H. Changes in prefrontal axons may disrupt the network in autism. J. Neurosci. 30, 14595–14609 (2010)
Chen, Y. et al. Variations in DNA elucidate molecular networks that cause disease. Nature 452, 429–435 (2008)
Plaisier, C. L. et al. A systems genetics approach implicates USF1, FADS3, and other causal candidate genes for familial combined hyperlipidemia. PLoS Genet. 5, e1000642 (2009)
Babatz, T. D., Kumar, R. A., Sudi, J., Dobyns, W. B. & Christian, S. L. Copy number and sequence variants implicate APBA2 as an autism candidate gene. Autism Res. 2, 359–364 (2009)
Castermans, D. et al. SCAMP5, NBEA and AMISYN: three candidate genes for autism involved in secretion of large dense-core vesicles. Hum. Mol. Genet. 19, 1368–1378 (2010)
Martin, C. L. et al. Cytogenetic and molecular characterization of A2BP1/FOX1 as a candidate gene for autism. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 144B, 869–876 (2007)
Alarcón, M. et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am. J. Hum. Genet. 82, 150–159 (2008)
Underwood, J. G., Boutz, P. L., Dougherty, J. D., Stoilov, P. & Black, D. L. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol. Cell. Biol. 25, 10005–10016 (2005)
Zhang, C. et al. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 22, 2550–2563 (2008)
Lee, J. A., Tang, Z. Z. & Black, D. L. An inducible change in Fox-1/A2BP1 splicing modulates the alternative splicing of downstream neuronal target exons. Genes Dev. 23, 2284–2293 (2009)
Moy, S. S., Nonneman, R. J., Young, N. B., Demyanenko, G. P. & Maness, P. F. Impaired sociability and cognitive function in Nrcam-null mice. Behav. Brain Res. 205, 123–131 (2009)
Zhao, X. et al. Significant association between the genetic variations in the 5′ end of the N-methyl-D-aspartate receptor subunit gene GRIN1 and schizophrenia. Biol. Psychiatry 59, 747–753 (2006)
Han, J. et al. A genome-wide association study identifies novel alleles associated with hair color and skin pigmentation. PLoS Genet. 4, e1000074 (2008)
Cooper, G. M. et al. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood 112, 1022–1027 (2008)
Morgan, J. T. et al. Microglial activation and increased microglial density observed in the dorsolateral prefrontal cortex in autism. Biol. Psychiatry 68, 368–376 (2010)
Boulanger, L. M. Immune proteins in brain development and synaptic plasticity. Neuron 64, 93–109 (2009)
Wang, K. et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature 459, 528–533 (2009)
Wintle, R. F. et al. (2010). A genotype resource for postmortem brain samples from the Autism Tissue Program. Autism Res. 4, 89–97 (2011)
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008)
Langfelder, P., Zhang, B. & Horvath, S. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics 24, 719–720 (2008)
Cahoy, J. D. et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 28, 264–278 (2008)
Albright, A. V. & Gonzalez-Scarano, F. Microarray analysis of activated mixed glial (microglia) and monocyte-derived macrophage gene expression. J. Neuroimmunol. 157, 27–38 (2004)
Oldham, M. C. et al. Functional organization of the transcriptome in human brain. Nature Neurosci. 11, 1271–1282 (2008)
Miller, J. A., Horvath, S. & Geschwind, D. H. Divergence of human and mouse brain transcriptome highlights Alzheimer disease pathways. Proc. Natl Acad. Sci. USA 107, 12698–12703 (2010)
Luco, R. F. et al. Regulation of alternative splicing by histone modifications. Science 327, 996–1000 (2010)
Pan, Q., Shai, O., Lee, L. J., Frey, B. J. & Blencowe, B. J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nature Genet. 40, 1413–1415 (2008)
Wang, K., Li, M. & Bucan, M. Pathway-based approaches for analysis of genomewide association studies. Am. J. Hum. Genet. 81, 1278–1283 (2007)
Zhang, K., Cui, S., Chang, S., Zhang, L. & Wang, J. i-GSEA4GWAS: a web server for identification of pathways/gene sets associated with traits by applying an improved gene set enrichment analysis to genome-wide association study. Nucleic Acids Res. 38 (suppl. 2). W90–W95 (2010)
Acknowledgements
We are grateful for the efforts of the Autism Tissue Program (ATP) of Autism Speaks and the families that have enrolled in the ATP, which made this work possible. We also thank S. Scherer and R. Wintle for sharing their SNP genotyping data on the AGP samples with us before its publication. We would also like to thank B. Abrahams for help in the initial stages of the project, B. Fogel, G. Konopka, N. Barbosa-Morais and J. Bomar for critically reading the manuscript, M. Lazaro for help with tissue dissection, and C. Vijayendran and K. Winden for useful discussions. This work was funded by an Autism Center of Excellence Network Grant from NIMH 5R01MH081754-03 and NIMH R37MH060233 to D.H.G. and by grants from the Canadian Institutes of Health Research and Genome Canada through the Ontario Genomics Institute to B.J.B. and others.
Author information
Authors and Affiliations
Contributions
I.V. and D.H.G. designed the study and wrote the manuscript. I.V. performed experiments, analysed the data and conducted the GWAS set enrichment analysis. X.W. and B.J.B. analysed the RNA sequencing data. J.K.L. contributed to the GWAS set enrichment analysis. Y.T. performed some of the microarray qRT-PCR validation experiments. R.M.C. supervised the GWAS set enrichment analysis. S.H. supervised the WGCNA analysis. P.J. and J.M. provided dissected tissue for the replication experiment. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
This file contains Supplementary Figures 1-9 with legends. (PDF 6856 kb)
Supplementary Tables
This file contains Supplementary Tables 1-14. (PDF 1146 kb)
Supplementary Data
This file contains Supplementary Data. (XLS 13307 kb)
Rights and permissions
About this article
Cite this article
Voineagu, I., Wang, X., Johnston, P. et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 474, 380–384 (2011). https://doi.org/10.1038/nature10110
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10110
This article is cited by
-
Identification of novel molecular subtypes to improve the classification framework of nasopharyngeal carcinoma
British Journal of Cancer (2024)
-
Remodeling of the postsynaptic proteome in male mice and marmosets during synapse development
Nature Communications (2024)
-
Prenatal Programming of Monocyte Chemotactic Protein-1 Signaling in Autism Susceptibility
Molecular Neurobiology (2024)
-
Novel Proline Transporter Inhibitor (LQFM215) Presents Antipsychotic Effect in Ketamine Model of Schizophrenia
Neurochemical Research (2024)
-
Tsc2 mutation rather than Tsc1 mutation dominantly causes a social deficit in a mouse model of tuberous sclerosis complex
Human Genomics (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.