Abstract
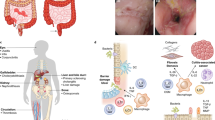
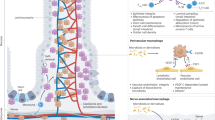
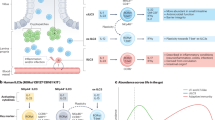
Intestinal homeostasis depends on complex interactions between the microbiota, the intestinal epithelium and the host immune system. Diverse regulatory mechanisms cooperate to maintain intestinal homeostasis, and a breakdown in these pathways may precipitate the chronic inflammatory pathology found in inflammatory bowel disease. It is now evident that immune effector modules that drive intestinal inflammation are conserved across innate and adaptive leukocytes and can be controlled by host regulatory cells. Recent evidence suggests that several factors may tip the balance between homeostasis and intestinal inflammation, presenting future challenges for the development of new therapies for inflammatory bowel disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Kaser, A., Zeissig, S. & Blumberg, R. S. Inflammatory bowel disease. Annu. Rev. Immunol. 28, 573–621 (2010).
Hill, D. A. & Artis, D. Intestinal bacteria and the regulation of immune cell homeostasis. Annu. Rev. Immunol. 28, 623–667 (2010).
Artis, D. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nature Rev. Immunol. 8, 411–420 (2008).
Hooper, L. V. & Macpherson, A. J. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nature Rev. Immunol. 10, 159–169 (2010).
Koslowski, M. J., Beisner, J., Stange, E. F. & Wehkamp, J. Innate antimicrobial host defense in small intestinal Crohn's disease. Int. J. Med. Microbiol. 300, 34–40 (2010).
Vaishnava, S., Behrendt, C. L., Ismail, A. S., Eckmann, L. & Hooper, L. V. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host–microbial interface. Proc. Natl Acad. Sci. USA 105, 20858–20863 (2008).
Cadwell, K. et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 456, 259–263 (2008).
Deretic, V. & Levine, B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe 5, 527–549 (2009).
Kaser, A., Martinez-Naves, E. & Blumberg, R. S. Endoplasmic reticulum stress: implications for inflammatory bowel disease pathogenesis. Curr. Opin. Gastroenterol. 26, 318–326 (2010).
Kaser, A. et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell 134, 743–756 (2008). This report links ER stress in IECs to the development of intestinal inflammation in both mice and humans.
Taylor, B. C. et al. TSLP regulates intestinal immunity and inflammation in mouse models of helminth infection and colitis. J. Exp. Med. 206, 655–667 (2009).
Abreu, M. T. Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nature Rev. Immunol. 10, 131–144 (2010).
Fagarasan, S., Kawamoto, S., Kanagawa, O. & Suzuki, K. Adaptive immune regulation in the gut: T cell-dependent and T cell-independent IgA synthesis. Annu. Rev. Immunol. 28, 243–273 (2010).
Slack, E. et al. Innate and adaptive immunity cooperate flexibly to maintain host–microbiota mutualism. Science 325, 617–620 (2009).
Asquith, M. J., Boulard, O., Powrie, F. & Maloy, K. J. Pathogenic and protective roles of MyD88 in leukocytes and epithelial cells in mouse models of inflammatory bowel disease. Gastroenterology 139, 519–529 (2010).
Cario, E. Toll-like receptors in inflammatory bowel diseases: a decade later. Inflamm. Bowel Dis. 16, 1583–1597 (2010).
Strober, W., Murray, P. J., Kitani, A. & Watanabe, T. Signalling pathways and molecular interactions of NOD1 and NOD2. Nature Rev. Immunol. 6, 9–20 (2006).
Chen, G. Y., Shaw, M. H., Redondo, G. & Nunez, G. The innate immune receptor Nod1 protects the intestine from inflammation-induced tumorigenesis. Cancer Res. 68, 10060–10067 (2008).
Dupaul-Chicoine, J. et al. Control of intestinal homeostasis, colitis, and colitis-associated colorectal cancer by the inflammatory caspases. Immunity 32, 367–378 (2010).
Zaki, M. H. et al. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity 32, 379–391 (2010).
Allen, I. C. et al. The NLRP3 inflammasome functions as a negative regulator of tumorigenesis during colitis-associated cancer. J. Exp. Med. 207, 1045–1056 (2010).
Schroder, K. & Tschopp, J. The inflammasomes. Cell 140, 821–832 (2010).
Siegmund, B. Interleukin-18 in intestinal inflammation: friend and foe? Immunity 32, 300–302 (2010).
Salcedo, R. et al. MyD88-mediated signaling prevents development of adenocarcinomas of the colon: role of interleukin 18. J. Exp. Med. 207, 1625–1636 (2010).
Feng, T., Wang, L., Schoeb, T. R., Elson, C. O. & Cong, Y. Microbiota innate stimulation is a prerequisite for T cell spontaneous proliferation and induction of experimental colitis. J. Exp. Med. 207, 1321–1332 (2010).
Asquith, M. & Powrie, F. An innately dangerous balancing act: intestinal homeostasis, inflammation, and colitis-associated cancer. J. Exp. Med. 207, 1573–1577 (2010).
Saleh, M. & Trinchieri, G. Innate immune mechanisms of colitis and colitis-associated colorectal cancer. Nature Rev. Immunol. 11, 9–20 (2011).
Round, J. L. & Mazmanian, S. K. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl Acad. Sci. USA 107, 12204–12209 (2010).
Richardson, W. M. et al. Nucleotide-binding oligomerization domain-2 inhibits Toll-like receptor-4 signaling in the intestinal epithelium. Gastroenterology 139, 904–917 (2010).
Travassos, L. H. et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nature Immunol. 11, 55–62 (2010).
Cooney, R. et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nature Med. 16, 90–97 (2010).
Saitoh, T. et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature 456, 264–268 (2008). References 31 and 32 describe a link between key Crohn's disease susceptibility factors by showing that NOD2 can stimulate autophagy and that this constitutes an important bacterial handling mechanism.
Zhou, R., Yazdi, A. S., Menu, P. & Tschopp, J. A role for mitochondria in NLRP3 inflammasome activation. Nature 469, 221–225 (2011).
Martinon, F., Chen, X., Lee, A. H. & Glimcher, L. H. TLR activation of the transcription factor XBP1 regulates innate immune responses in macrophages. Nature Immunol. 11, 411–418 (2010).
Melmed, G. Y. & Targan, S. R. Future biologic targets for IBD: potentials and pitfalls. Nature Rev. Gastroenterol. Hepatol. 7, 110–117 (2010).
Maloy, K. J. & Kullberg, M. C. IL-23 and Th17 cytokines in intestinal homeostasis. Mucosal Immunol. 1, 339–349 (2008).
Kamada, N. et al. Unique CD14+ intestinal macrophages contribute to the pathogenesis of Crohn disease via IL-23/IFN-γ axis. J. Clin. Invest. 118, 2269–2280 (2008).
Goodall, J. C. et al. Endoplasmic reticulum stress-induced transcription factor, CHOP, is crucial for dendritic cell IL-23 expression. Proc. Natl Acad. Sci. USA 107, 17698–17703 (2010).
Ahern, P. P. et al. Interleukin-23 drives intestinal inflammation through direct activity on T cells. Immunity 33, 279–288 (2010).
Leppkes, M. et al. RORγ-expressing Th17 cells induce murine chronic intestinal inflammation via redundant effects of IL-17A and IL-17F. Gastroenterology 136, 257–267 (2009).
Cosmi, L. et al. Human interleukin 17-producing cells originate from a CD161+CD4+ T cell precursor. J. Exp. Med. 205, 1903–1916 (2008).
Kleinschek, M. A. et al. Circulating and gut-resident human Th17 cells express CD161 and promote intestinal inflammation. J. Exp. Med. 206, 525–534 (2009).
Yang, X. O. et al. Regulation of inflammatory responses by IL-17F. J. Exp. Med. 205, 1063–1075 (2008).
Chaudhry, A. et al. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science 326, 986–991 (2009).
Buonocore, S. et al. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature 464, 1371–1375 (2010).
Wu, S. et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nature Med. 15, 1016–1022 (2009).
Ivanov, I. I. et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009).
Gaboriau-Routhiau, V. et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31, 677–689 (2009). References 47 and 48 document a strong link between segmented filamentous bacteria colonization and T H 17 cells and, together with reference 80, provide compelling evidence that colonization with distinct types of commensal bacterium results in the accumulation of different effector T cells in the intestine.
Wolk, K., Witte, E., Witte, K., Warszawska, K. & Sabat, R. Biology of interleukin-22. Semin. Immunopathol. 32, 17–31 (2010).
Pickert, G. et al. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J. Exp. Med. 206, 1465–1472 (2009).
Sonnenberg, G. F. et al. Pathological versus protective functions of IL-22 in airway inflammation are regulated by IL-17A. J. Exp. Med. 207, 1293–1305 (2010).
Chung, Y. et al. Critical regulation of early Th17 cell differentiation by interleukin-1 signaling. Immunity 30, 576–587 (2009).
Ng, J. et al. Clostridium difficile toxin-induced inflammation and intestinal injury are mediated by the inflammasome. Gastroenterology 139, 542–552.e3 (2010).
Muller, A. J. et al. The S. Typhimurium effector SopE induces caspase-1 activation in stromal cells to initiate gut inflammation. Cell Host Microbe 6, 125–136 (2009).
Colonna, M. Interleukin-22-producing natural killer cells and lymphoid tissue inducer-like cells in mucosal immunity. Immunity 31, 15–23 (2009).
Cua, D. J. & Tato, C. M. Innate IL-17-producing cells: the sentinels of the immune system. Nature Rev. Immunol. 10, 479–489 (2010).
Park, S. G. et al. T regulatory cells maintain intestinal homeostasis by suppressing γδ T cells. Immunity 33, 791–803 (2010).
Martin, B., Hirota, K., Cua, D. J., Stockinger, B. & Veldhoen, M. Interleukin-17-producing γδ T cells selectively expand in response to pathogen products and environmental signals. Immunity 31, 321–330 (2009).
Sutton, C. E. et al. Interleukin-1 and IL-23 induce innate IL-17 production from γδ T cells, amplifying Th17 responses and autoimmunity. Immunity 31, 331–341 (2009).
Spits, H. & Di Santo, J. P. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nature Immunol. 12, 21–27 (2011).
Sawa, S. et al. Lineage relationship analysis of RORγ+ innate lymphoid cells. Science 330, 665–669 (2010).
Sonnenberg, G. F., Monticelli, L. A., Elloso, M. M., Fouser, L. A. & Artis, D. CD4+ lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity 34, 122–134 (2011). References 45 and 62 identify new populations of LTI-like ILCs that secrete T H 1 and T H 17 pro-inflammatory cytokines in response to IL-23, and these contribute to intestinal pathology and host defences against intestinal pathogenic bacteria.
Moro, K. et al. Innate production of TH2 cytokines by adipose tissue-associated c-Kit+Sca-1+ lymphoid cells. Nature 463, 540–544 (2010).
Saenz, S. A. et al. IL25 elicits a multipotent progenitor cell population that promotes TH2 cytokine responses. Nature 464, 1362–1366 (2010).
Neill, D. R. et al. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature 464, 1367–1370 (2010). References 63–65 describe various ILC populations that secrete T H 2 cytokines (ILC type 2) in response to IL-25 and IL-33 and that can contribute to defence against intestinal helminth infection.
Coombes, J. L. & Powrie, F. Dendritic cells in intestinal immune regulation. Nature Rev. Immunol. 8, 435–446 (2008).
Varol, C., Zigmond, E. & Jung, S. Securing the immune tightrope: mononuclear phagocytes in the intestinal lamina propria. Nature Rev. Immunol. 10, 415–426 (2010).
Varol, C. et al. Intestinal lamina propria dendritic cell subsets have different origin and functions. Immunity 31, 502–512 (2009).
Bogunovic, M. et al. Origin of the lamina propria dendritic cell network. Immunity 31, 513–525 (2009).
Schulz, O. et al. Intestinal CD103+, but not CX3CR1+, antigen sampling cells migrate in lymph and serve classical dendritic cell functions. J. Exp. Med. 206, 3101–3114 (2009).
Niess, J. H. & Adler, G. Enteric flora expands gut lamina propria CX3CR1+ dendritic cells supporting inflammatory immune responses under normal and inflammatory conditions. J. Immunol. 184, 2026–2037 (2010).
Manicassamy, S. et al. Activation of β-catenin in dendritic cells regulates immunity versus tolerance in the intestine. Science 329, 849–853 (2010).
Laffont, S., Siddiqui, K. R. & Powrie, F. Intestinal inflammation abrogates the tolerogenic properties of MLN CD103+ dendritic cells. Eur. J. Immunol. 40, 1877–1883 (2010).
Smith, P. D. et al. Intestinal macrophages and response to microbial encroachment. Mucosal Immunol. 4, 31–42 (2011).
Murai, M. et al. Interleukin 10 acts on regulatory T cells to maintain expression of the transcription factor Foxp3 and suppressive function in mice with colitis. Nature Immunol. 10, 1178–1184 (2009).
Hadis, U. et al. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity 34, 237–246 (2011). This study shows that oral tolerance requires homing and expansion of T reg cells in the intestine.
Siddiqui, K. R., Laffont, S. & Powrie, F. E-cadherin marks a subset of inflammatory dendritic cells that promote T cell-mediated colitis. Immunity 32, 557–567 (2010).
Platt, A. M., Bain, C. C., Bordon, Y., Sester, D. P. & Mowat, A. M. An independent subset of TLR expressing CCR2-dependent macrophages promotes colonic inflammation. J. Immunol. 184, 6843–6854 (2010).
Izcue, A., Coombes, J. L. & Powrie, F. Regulatory lymphocytes and intestinal inflammation. Annu. Rev. Immunol. 27, 313–338 (2009).
Atarashi, K. et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331, 337–341 (2011). This paper describes a link between Clostridium spp. and T reg cells in the gut and, together with references 47 and 48, provides compelling evidence that colonization with distinct commensal bacteria results in the accumulation of different effector T cells in the intestine.
Littman, D. R. & Rudensky, A. Y. Th17 and regulatory T cells in mediating and restraining inflammation. Cell 140, 845–858 (2010).
Hall, J. A. et al. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity 29, 637–649 (2008).
Griseri, T., Asquith, M., Thompson, C. & Powrie, F. OX40 is required for regulatory T cell-mediated control of colitis. J. Exp. Med. 207, 699–709 (2010).
Oldenhove, G. et al. Decrease of Foxp3+ Treg cell number and acquisition of effector cell phenotype during lethal infection. Immunity 31, 772–786 (2009).
Li, M. O. & Flavell, R. A. Contextual regulation of inflammation: a duet by transforming growth factor-β and interleukin-10. Immunity 28, 468–476 (2008).
Fantini, M. C. et al. Smad7 controls resistance of colitogenic T cells to regulatory T cell-mediated suppression. Gastroenterology 136, 1308–1316.e3 (2009).
Pesu, M. et al. T-cell-expressed proprotein convertase furin is essential for maintenance of peripheral immune tolerance. Nature 455, 246–250 (2008).
Perruche, S. et al. CD3-specific antibody-induced immune tolerance involves transforming growth factor-β from phagocytes digesting apoptotic T cells. Nature Med. 14, 528–535 (2008).
Torchinsky, M. B., Garaude, J., Martin, A. P. & Blander, J. M. Innate immune recognition of infected apoptotic cells directs TH17 cell differentiation. Nature 458, 78–82 (2009).
Cong, Y., Feng, T., Fujihashi, K., Schoeb, T. R. & Elson, C. O. A dominant, coordinated T regulatory cell–IgA response to the intestinal microbiota. Proc. Natl Acad. Sci. USA 106, 19256–19261 (2009).
Saraiva, M. & O'Garra, A. The regulation of IL-10 production by immune cells. Nature Rev. Immunol. 10, 170–181 (2010).
Glocker, E. O. et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N. Engl. J. Med. 361, 2033–2045 (2009).
Smith, A. M. et al. Disordered macrophage cytokine secretion underlies impaired acute inflammation and bacterial clearance in Crohn's disease. J. Exp. Med. 206, 1883–1897 (2009).
Winter, S. E. et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429 (2010).
Sartor, R. B. Microbial influences in inflammatory bowel diseases. Gastroenterology 134, 577–594 (2008).
Frank, D. N. et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm. Bowel Dis. 17, 179–184 (2011).
Willing, B. P. et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 139, 1844–1854.e1 (2010).
Maslowski, K. M. & Mackay, C. R. Diet, gut microbiota and immune responses. Nature Immunol. 12, 5–9 (2011).
Cadwell, K. et al. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell 141, 1135–1145 (2010).
Garrett, W. S. et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe 8, 292–300 (2010). References 99 and 100 provide illustrative examples of how several host and environmental factors may act together to precipitate chronic intestinal inflammation.
Acknowledgements
We thank M. Asquith, H. Uhlig, P. Ahern and M. Barnes for review and G. Song-Zhao and O. Harrison for help with the figures. We apologize to those whose work was not cited owing to space constraints. K.J.M. and F.P. are supported by grants from the Wellcome Trust, Cancer Research UK and the European Union (FP7, INFLAMMACARE).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reprints and permissions information is available at http://www.nature.com/reprints.
Supplementary information
Supplementary References
This file contains a Supplementary Bibliography to the main paper. (PDF 261 kb)
Rights and permissions
About this article
Cite this article
Maloy, K., Powrie, F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature 474, 298–306 (2011). https://doi.org/10.1038/nature10208
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10208
This article is cited by
-
Advancing the frontiers of colorectal cancer treatment: harnessing ferroptosis regulation
Apoptosis (2024)
-
Effects of SCFAs and TMAO on non-alcoholic fatty liver disease indicating the therapeutic benefits of plant-based diet, and supplemental prebiotics, probiotics and synbiotics
Applied Biological Chemistry (2023)
-
Akkermansia muciniphila and its membrane protein ameliorates intestinal inflammatory stress and promotes epithelial wound healing via CREBH and miR-143/145
Journal of Biomedical Science (2023)
-
Role of Rho GTPases in inflammatory bowel disease
Cell Death Discovery (2023)
-
Optical imaging of the small intestine immune compartment across scales
Communications Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.