Abstract
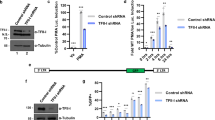
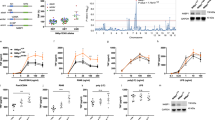
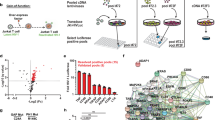
Interferon regulatory factor 4 (IRF4) is an IRF family transcription factor with critical roles in lymphoid development and in regulating the immune response1,2. IRF4 binds DNA weakly owing to a carboxy-terminal auto-inhibitory domain, but cooperative binding with factors such as PU.1 or SPIB in B cells increases binding affinity3, allowing IRF4 to regulate genes containing ETS–IRF composite elements (EICEs; 5′-GGAAnnGAAA-3′)1. Here we show that in mouse CD4+ T cells, where PU.1/SPIB expression is low, and in B cells, where PU.1 is well expressed, IRF4 unexpectedly can cooperate with activator protein-1 (AP1) complexes to bind to AP1–IRF4 composite (5′-TGAnTCA/GAAA-3′) motifs that we denote as AP1–IRF composite elements (AICEs). Moreover, BATF–JUN family protein complexes cooperate with IRF4 in binding to AICEs in pre-activated CD4+ T cells stimulated with IL-21 and in TH17 differentiated cells. Importantly, BATF binding was diminished in Irf4−/− T cells and IRF4 binding was diminished in Batf−/− T cells, consistent with functional cooperation between these factors. Moreover, we show that AP1 and IRF complexes cooperatively promote transcription of the Il10 gene, which is expressed in TH17 cells and potently regulated by IL-21. These findings reveal that IRF4 can signal via complexes containing ETS or AP1 motifs depending on the cellular context, thus indicating new approaches for modulating IRF4-dependent transcription.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Taniguchi, T., Ogasawara, K., Takaoka, A. & Tanaka, N. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19, 623–655 (2001)
Lohoff, M. & Mak, T. W. Roles of interferon-regulatory factors in T-helper-cell differentiation. Nature Rev. Immunol. 5, 125–135 (2005)
Brass, A. L., Kehrli, E., Eisenbeis, C. F., Storb, U. & Singh, H. Pip, a lymphoid-restricted IRF, contains a regulatory domain that is important for autoinhibition and ternary complex formation with the Ets factor PU.1. Genes Dev. 10, 2335–2347 (1996)
Grossman, A. et al. Cloning of human lymphocyte-specific interferon regulatory factor (hLSIRF/hIRF4) and mapping of the gene to 6p23–p25. Genomics 37, 229–233 (1996)
Grumont, R. J. & Gerondakis, S. Rel induces interferon regulatory factor 4 (IRF-4) expression in lymphocytes: modulation of interferon-regulated gene expression by Rel/nuclear factor κB. J. Exp. Med. 191, 1281–1292 (2000)
Escalante, C. R. et al. Crystal structure of PU.1/IRF-4/DNA ternary complex. Mol. Cell 10, 1097–1105 (2002)
Pernis, A. B. The role of IRF-4 in B and T cell activation and differentiation. J. Interferon Cytokine Res. 22, 111–120 (2002)
Lohoff, M. et al. Dysregulated T helper cell differentiation in the absence of interferon regulatory factor 4. Proc. Natl Acad. Sci. USA 99, 11808–11812 (2002)
Tominaga, N. et al. Development of Th1 and not Th2 immune responses in mice lacking IFN-regulatory factor-4. Int. Immunol. 15, 1–10 (2003)
Rengarajan, J. et al. Interferon regulatory factor 4 (IRF4) interacts with NFATc2 to modulate interleukin 4 gene expression. J. Exp. Med. 195, 1003–1012 (2002)
Hu, C. M., Jang, S. Y., Fanzo, J. C. & Pernis, A. B. Modulation of T cell cytokine production by interferon regulatory factor-4. J. Biol. Chem. 277, 49238–49246 (2002)
Staudt, V. et al. Interferon-regulatory factor 4 is essential for the developmental program of T helper 9 cells. Immunity 33, 192–202 (2010)
Brüstle, A. et al. The development of inflammatory TH-17 cells requires interferon-regulatory factor 4. Nature Immunol. 8, 958–966 (2007)
Kwon, H. et al. Analysis of interleukin-21-induced Prdm1 gene regulation reveals functional cooperation of STAT3 and IRF4 transcription factors. Immunity 31, 941–952 (2009)
Schraml, B. U. et al. The AP-1 transcription factor Batf controls TH17 differentiation. Nature 460, 405–409 (2009)
Echlin, D. R., Tae, H. J., Mitin, N. & Taparowsky, E. J. B-ATF functions as a negative regulator of AP-1 mediated transcription and blocks cellular transformation by Ras and Fos. Oncogene 19, 1752–1763 (2000)
Dorsey, M. J. et al. B-ATF: a novel human bZIP protein that associates with members of the AP-1 transcription factor family. Oncogene 11, 2255–2265 (1995)
Korn, T. et al. IL-21 initiates an alternative pathway to induce proinflammatory TH17 cells. Nature 448, 484–487 (2007)
Nurieva, R. et al. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature 448, 480–483 (2007)
Zhou, L. et al. IL-6 programs TH-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nature Immunol. 8, 967–974 (2007)
Zhang, X., Wrzeszczynska, M. H., Horvath, C. M. & Darnell, J. E., Jr Interacting regions in Stat3 and c-Jun that participate in cooperative transcriptional activation. Mol. Cell. Biol. 19, 7138–7146 (1999)
Spolski, R., Kim, H. P., Zhu, W., Levy, D. E. & Leonard, W. J. IL-21 mediates suppressive effects via its induction of IL-10. J. Immunol. 182, 2859–2867 (2009)
Pot, C. et al. Cutting edge: IL-27 induces the transcription factor c-Maf, cytokine IL-21, and the costimulatory receptor ICOS that coordinately act together to promote differentiation of IL-10-producing Tr1 cells. J. Immunol. 183, 797–801 (2009)
Lee, C. G. et al. A distal cis-regulatory element, CNS-9, controls NFAT1 and IRF4-mediated IL-10 gene activation in T helper cells. Mol. Immunol. 46, 613–621 (2009)
Chang, H. C. et al. PU.1 expression delineates heterogeneity in primary Th2 cells. Immunity 22, 693–703 (2005)
Chang, H. C. et al. The transcription factor PU.1 is required for the development of IL-9-producing T cells and allergic inflammation. Nature Immunol. 11, 527–534 (2010)
Altin, J. A., Goodnow, C. C. & Cook, M. C. IL-10+CTLA-4+ Th2 inhibitory cells form in a Foxp3-independent, IL-2-dependent manner from Th2 effectors during chronic inflammation. J. Immunol. 188, 5478–5488 (2012)
Tussiwand, R. et al. Compensatory dendritic cell development mediated by BATF–IRF interactions. Nature https://doi.org/10.1038/nature11531 (19 September 2012)
Liao, W., Lin, J. X., Wang, L., Li, P. & Leonard, W. J. Modulation of cytokine receptors by IL-2 broadly regulates differentiation into helper T cell lineages. Nature Immunol. 12, 551–559 (2011)
Lin, J. X. et al. Critical role of STAT5 transcription factor tetramerization for cytokine responses and normal immune function. Immunity 36, 586–599 (2012)
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009)
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008)
Bailey, T. L. & Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36 (1994)
Acknowledgements
This work was supported by the Division of Intramural Research, National Heart, Lung, and Blood Institute, NIH (P.L., R.S., W.L., L.W. and W.J.L.) and the Howard Hughes Medical Institute (T.L.M. and K.M.M.). We thank J.-X. Lin for valuable suggestions, critical comments and RNA-Seq data for pro-B/pre-B-enriched populations. We thank J. Zhu and Y. Wakabayashi, NHLBI DNA Sequencing Core, for excellent services, K. Ozato and Y. Hiroaki, NICHD, for Irf4−/− mice, and J. Thierry-Mieg and D. Thierry-Mieg, NCBI, for early analysis of ChIP-Seq data from ref. 14.
Author information
Authors and Affiliations
Contributions
P.L. designed experiments, analysed data and wrote the paper. R.S. designed and performed experiments, analysed data and wrote the paper. W.L. and L.W. designed and performed experiments and analysed data. T.L.M. and K.M.M. provided reagents and made suggestions. W.J.L. designed experiments, analysed data and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
W.J.L. and R.S. are inventors on NIH patents related to IL-21.
Additional information
Data sets (ChIP-Seq and RNA-Seq data) have been deposited in the Gene Expression Omnibus (GSE39756).
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-8 and Supplementary Tables 1-2. (PDF 1876 kb)
Rights and permissions
About this article
Cite this article
Li, P., Spolski, R., Liao, W. et al. BATF–JUN is critical for IRF4-mediated transcription in T cells. Nature 490, 543–546 (2012). https://doi.org/10.1038/nature11530
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11530
This article is cited by
-
BATF and BATF3 deficiency alters CD8+ effector/exhausted T cells balance in skin transplantation
Molecular Medicine (2024)
-
CD4+ T cell immunity is dependent on an intrinsic stem-like program
Nature Immunology (2024)
-
IKAROS and AIOLOS directly regulate AP-1 transcriptional complexes and are essential for NK cell development
Nature Immunology (2024)
-
Cardinal features of immune memory in innate lymphocytes
Nature Immunology (2023)
-
The association of immune-related genes and the potential role of IL10 with biliary atresia
Pediatric Research (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.