Abstract
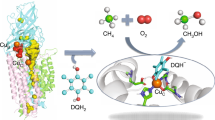
Methanotrophs consume methane as their major carbon source and have an essential role in the global carbon cycle by limiting escape of this greenhouse gas to the atmosphere1,2,3. These bacteria oxidize methane to methanol by soluble and particulate methane monooxygenases (MMOs)1,2,3,4. Soluble MMO contains three protein components, a 251-kilodalton hydroxylase (MMOH), a 38.6-kilodalton reductase (MMOR), and a 15.9-kilodalton regulatory protein (MMOB), required to couple electron consumption with substrate hydroxylation at the catalytic diiron centre of MMOH2. Until now, the role of MMOB has remained ambiguous owing to a lack of atomic-level information about the MMOH–MMOB (hereafter termed H–B) complex. Here we remedy this deficiency by providing a crystal structure of H–B, which reveals the manner by which MMOB controls the conformation of residues in MMOH crucial for substrate access to the active site. MMOB docks at the α2β2 interface of α2β2γ2 MMOH, and triggers simultaneous conformational changes in the α-subunit that modulate oxygen and methane access as well as proton delivery to the diiron centre. Without such careful control by MMOB of these substrate routes to the diiron active site, the enzyme operates as an NADH oxidase rather than a monooxygenase5. Biological catalysis involving small substrates is often accomplished in nature by large proteins and protein complexes. The structure presented in this work provides an elegant example of this principle.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hanson, R. S. & Hanson, T. E. Methanotrophic bacteria. Microbiol. Rev. 60, 439–471 (1996)
Merkx, M. et al. Dioxygen activation and methane hydroxylation by soluble methane monooxygenase: A tale of two irons and three proteins. Angew. Chem. Int. Ed. Engl. 40, 2782–2807 (2001)
Wallar, B. J. & Lipscomb, J. D. Dioxygen activation by enzymes containing binuclear non-heme iron clusters. Chem. Rev. 96, 2625–2658 (1996)
Leahy, J. G., Batchelor, P. J. & Morcomb, S. M. Evolution of the soluble diiron monooxygenases. FEMS Microbiol. Rev. 27, 449–479 (2003)
Gassner, G. T. & Lippard, S. J. Component interactions in the soluble methane monooxygenase system from Methylococcus capsulatus (Bath). Biochemistry 38, 12768–12785 (1999)
Elango, N. et al. Crystal structure of the hydroxylase component of methane monooxygenase from Methylosinus trichosporium OB3b. Protein Sci. 6, 556–568 (1997)
Rosenzweig, A. C., Frederick, C. A., Lippard, S. J. & Nordlund, P. Crystal structure of a bacterial non-haem iron hydroxylase that catalyzes the biological oxidation of methane. Nature 366, 537–543 (1993)
Shu, L. et al. An Fe2 IVO2 diamond core structure for the key intermediate Q of methane monooxygenase. Science 275, 515–518 (1997)
Tinberg, C. E. & Lippard, S. J. Revisiting the mechanism of dioxygen activation in soluble methane monooxygenase from M. capsulatus (Bath): Evidence for a multi-step, proton-dependent reaction pathway. Biochemistry 48, 12145–12158 (2009)
Whittington, D. A. & Lippard, S. J. Crystal structures of the soluble methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath) demonstrating geometrical variability at the dinuclear iron active site. J. Am. Chem. Soc. 123, 827–838 (2001)
Paulsen, K. E. et al. Oxidation-reduction potentials of the methane monooxygenase hydroxylase component from Methylosinus trichosporium OB3b. Biochemistry 33, 713–722 (1994)
Froland, W. A., Andersson, K. K., Lee, S. K., Liu, Y. & Lipscomb, J. D. Methane monooxygenase component B and reductase alter the regioselectivity of the hydroxylase component-catalyzed reactions. J. Biol. Chem. 267, 17588–17597 (1992)
Liu, K. E. et al. Kinetic and spectroscopic characterization of intermediates and component interactions in reactions of methane monooxygenase from Methylococcus capsulatus (Bath). J. Am. Chem. Soc. 117, 10174–10185 (1995)
Liu, Y., Nesheim, J. C., Lee, S. K. & Lipscomb, J. D. Gating effects of component B on oxygen activation by the methane monooxygenase hydroxylase component. J. Biol. Chem. 270, 24662–24665 (1995)
Bailey, L. J., Mccoy, J. G., Phillips, G. N. & Fox, B. G. Structural consequences of effector protein complex formation in a diiron hydroxylase. Proc. Natl Acad. Sci. USA 105, 19194–19198 (2008)
Sazinsky, M. H. & Lippard, S. J. Correlating structure with function in bacterial multicomponent monooxygenases and related diiron proteins. Acc. Chem. Res. 39, 558–566 (2006)
Sazinsky, M. H., Dunten, P. W., McCormick, M. S., DiDonato, A. & Lippard, S. J. X-ray structure of a hydroxylase-regulatory protein complex from a hydrocarbon-oxidizing multicomponent monooxygenase, Pseudomonas sp. OX1 phenol hydroxylase. Biochemistry 45, 15392–15404 (2006)
Walters, K. J., Gassner, G. T., Lippard, S. J. & Wagner, G. Structure of the soluble methane monooxygenase regulatory protein B. Proc. Natl Acad. Sci. USA 96, 7877–7882 (1999)
Brandstetter, H., Whittington, D. A., Lippard, S. J. & Frederick, C. A. Mutational and structural analyses of the regulatory protein B of soluble methane monooxygenase from Methylococcus capsulatus (Bath). Chem. Biol. 6, 441–449 (1999)
Chang, S. L., Wallar, B. J., Lipscomb, J. D. & Mayo, K. H. Residues in Methylosinus trichosporium OB3b methane monooxygenase component B involved in molecular interactions with reduced- and oxidized-hydroxylase component: a role for the N-terminus. Biochemistry 40, 9539–9551 (2001)
Blazyk, J. L., Gassner, G. T. & Lippard, S. J. Intermolecular electron-transfer reactions in soluble methane monooxygenase: A role for hysteresis in protein function. J. Am. Chem. Soc. 127, 17364–17376 (2005)
McCormick, M. S. & Lippard, S. J. Analysis of substrate access to active sites in bacterial multicomponent monooxygenase hydroxylases: X-ray crystal structure of xenon-pressurized phenol hydroxylase from Pseudomonas sp OX1. Biochemistry 50, 11058–11069 (2011)
Lee, S. K. & Lipscomb, J. D. Oxygen activation catalyzed by methane monooxygenase hydroxylase component: proton delivery during the O-O bond cleavage steps. Biochemistry 38, 4423–4432 (1999)
Song, W. J. et al. Active site threonine facilitates proton transfer during dioxygen activation at the diiron center of toluene/o-xylene monooxygenase hydroxylase. J. Am. Chem. Soc. 132, 13582–13585 (2010)
Rardin, R. L., Tolman, W. B. & Lippard, S. J. Monodentate carboxylate complexes and the carboxylate shift: Implications for polymetalloprotein structure and function. New J. Chem. 15, 417–430 (1991)
Whittington, D. A., Rosenzweig, A. C., Frederick, C. A. & Lippard, S. J. Xenon and halogenated alkanes track putative substrate binding cavities in the soluble methane monooxygenase hydroxylase. Biochemistry 40, 3476–3482 (2001)
Whittington, D. A., Sazinsky, M. H. & Lippard, S. J. X-ray crystal structure of alcohol products bound at the active site of soluble methane monooxygenase hydroxylase. J. Am. Chem. Soc. 123, 1794–1795 (2001)
Rosenzweig, A. C. et al. Crystal structures of the methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath): Implications for substrate gating and component interactions. Proteins 29, 141–152 (1997)
Coufal, D. E. et al. Sequencing and analysis of the Methylococcus capsulatus (Bath) soluble methane monooxygenase genes. Eur. J. Biochem. 267, 2174–2185 (2000)
Valentine, A. M., Stahl, S. S. & Lippard, S. J. Mechanistic studies of the reaction of reduced methane monooxygenase hydroxylase with dioxygen and substrates. J. Am. Chem. Soc. 121, 3876–3887 (1999)
Beauvais, L. G. & Lippard, S. J. Reactions of the peroxo intermediate of soluble methane monooxygenase hydroxylase with ethers. J. Am. Chem. Soc. 127, 7370–7378 (2005)
Liu, K. E., Johnson, C. C., Newcomb, M. & Lippard, S. J. Radical clock substrate probes and kinetic isotope effect studies of the hydroxylation of hydrocarbons by methane monooxygenase. J. Am. Chem. Soc. 115, 939–947 (1993)
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010)
Evans, P. Scaling and assessment of data quality. Acta Crystallogr. D 62, 72–82 (2006)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Acknowledgements
This work was supported by grant GM 32114 from the National Institute of General Medical Sciences to S. J. Lippard. We thank the staff at the Advanced Light Source beamline 8.2.2. in Lawrence Berkeley National Laboratory for the data collection, S. C. Harrison for resources and comments on the manuscript, and T. C. Johnstone, A. D. Liang and T.-T. Lu for discussions.
Author information
Authors and Affiliations
Contributions
S. J. Lee designed experiments, purified proteins and measured the enzyme activity, analysed data, and wrote the manuscript; M.S.M. analysed the enzyme cavity, analysed data, prepared figures, and wrote the manuscript; S. J. Lippard directed the project, designed experiments, analysed data, and wrote the manuscript; and U.-S.C. obtained crystals, solved and refined the structures, analysed data, and wrote the manuscript. All authors discussed the results and commented on the manuscripts.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-2, Supplementary Figures 1-12 and additional references. (PDF 1559 kb)
Rights and permissions
About this article
Cite this article
Lee, S., McCormick, M., Lippard, S. et al. Control of substrate access to the active site in methane monooxygenase. Nature 494, 380–384 (2013). https://doi.org/10.1038/nature11880
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11880
This article is cited by
-
Protein interface redesign facilitates the transformation of nanocage building blocks to 1D and 2D nanomaterials
Nature Communications (2021)
-
Bimetallic cooperation across the periodic table
Nature Reviews Chemistry (2020)
-
Misconceptions and challenges in methane-to-methanol over transition-metal-exchanged zeolites
Nature Catalysis (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.