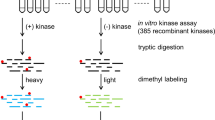
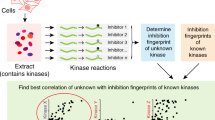
Abstract
Constitutive activation of one or more kinase signaling pathways is a hallmark of many cancers. Here we extend the previously described mass spectrometry–based KAYAK approach by monitoring kinase activities from multiple signaling pathways simultaneously. This improved single-reaction strategy, which quantifies the phosphorylation of 90 synthetic peptides in a single mass spectrometry run, is compatible with nanogram to microgram amounts of cell lysate. Furthermore, the approach enhances kinase monospecificity through substrate competition effects, faithfully reporting the signatures of many signaling pathways after mitogen stimulation or of basal pathway activation differences across a panel of well-studied cancer cell lines. Hierarchical clustering of activities from related experiments groups peptides phosphorylated by similar kinases together and, when combined with pathway alteration using pharmacological inhibitors, distinguishes underlying differences in potency, off-target effects and genetic backgrounds. Finally, we introduce a strategy to identify the kinase, and even associated protein complex members, responsible for phosphorylation events of interest.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Krause, D.S. & Van Etten, R.A. Tyrosine kinases as targets for cancer therapy. N. Engl. J. Med. 353, 172–187 (2005).
Sebolt-Leopold, J.S. & English, J.M. Mechanisms of drug inhibition of signalling molecules. Nature 441, 457–462 (2006).
Diks, S.H. et al. Kinome profiling for studying lipopolysaccharide signal transduction in human peripheral blood mononuclear cells. J. Biol. Chem. 279, 49206–49213 (2004).
Houseman, B.T., Huh, J.H., Kron, S.J. & Mrksich, M. Peptide chips for the quantitative evaluation of protein kinase activity. Nat. Biotechnol. 20, 270–274 (2002).
Janes, K.A. et al. A high-throughput quantitative multiplex kinase assay for monitoring information flow in signaling networks: application to sepsis-apoptosis. Mol. Cell. Proteomics 2, 463–473 (2003).
Shults, M.D. et al. A multiplexed protein kinase assay. ChemBioChem 8, 933–942 (2007).
Gao, H. & Leary, J.A. Multiplex inhibitor screening and kinetic constant determinations for yeast hexokinase using mass spectrometry based assays. J. Am. Soc. Mass Spectrom. 14, 173–181 (2003).
Pi, N., Armstrong, J.I., Bertozzi, C.R. & Leary, J.A. Kinetic analysis of NodST sulfotransferase using an electrospray ionization mass spectrometry assay. Biochemistry 41, 13283–13288 (2002).
Yu, Y. et al. A site-specific, multiplexed kinase activity assay using stable isotope dilution theory and high-resolution mass spectrometry. Proc. Natl. Acad. Sci. USA 106, 11606–11611 (2009).
Beausoleil, S.A. et al. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. USA 101, 12130–12135 (2004).
Villen, J., Beausoleil, S.A., Gerber, S.A. & Gygi, S.P. Large-scale phosphorylation analysis of mouse liver. Proc. Natl. Acad. Sci. USA 104, 1488–1493 (2007).
Manning, B.D. & Cantley, L.C. AKT/PKB signaling: navigating downstream. Cell 129, 1261–1274 (2007).
Ubersax, J.A. & Ferrell, J.E.J. Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 8, 530–541 (2007).
Alessi, D.R., Caudwell, F.B., Andjelkovic, M., Hemmings, B.A. & Cohen, P. Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 kinase. FEBS Lett. 399, 333–338 (1996).
Anjum, R. & Blenis, J. The RSK family of kinases: emerging roles in cellular signalling. Nat. Rev. Mol. Cell Biol. 9, 747–758 (2008).
Sigoillot, F.D., Sigoillot, S.M. & Guy, H.I. Breakdown of the regulatory control of pyrimidine biosynthesis in human breast cancer cells. Int. J. Cancer 109, 491–498 (2004).
Chou, T.F. et al. 31P NMR and genetic analysis establish hinT as the only Escherchia coli purine nucleoside phosphoramidase and as essential for growth under high salt conditions. J. Biol. Chem. 280, 15356–15361 (2005).
Astoul, E., Edmunds, C., Cantrell, D.A. & Ward, S.G. PI 3-K and T-cell activation: limitations of T-leukemic cell lines as signaling models. Trends Immunol. 22, 490–496 (2001).
Bain, J. et al. The selectivity of protein kinase inhibitors: a further update. Biochem. J. 408, 297–315 (2007).
Garcia-Echeverria, C. & Sellers, W.R. Drug discovery approaches targeting the PI3K/Akt pathway in cancer. Oncogene 27, 5511–5526 (2008).
Cheng, J.Q., Lindsley, C.W., Cheng, G.Z., Yang, H. & Nicosia, S.V. The Akt/PKB pathway: molecular target for cancer drug discovery. Oncogene 24, 7482–7492 (2005).
Peifer, C. & Alessi, D.R. Small-molecule inhibitors of PDK1. ChemMedChem 3, 1810–1838 (2008).
Xu, Z. et al. Development of high throughput HTRF and Alphascreen assays for identification of potent inhibitors of PDK1. J. Biomol. Screen. (in the press) (2009).
Sun, D. et al. Efficient identification of novel leads by dynamic focused screening: PDK1 case study. Comb. Chem. High Throughput Screen. (in the press) (2009).
Bilodeau, M.T. et al. Allosteric inhibitors of Akt1 and Akt2: a naphthyridinone with efficacy in an A2780 tumor xenograft model. Bioorg. Med. Chem. Lett. 18, 3178–3182 (2008).
Manning, G., Whyte, D.B., Martinez, R., Hunter, T. & Sudarsanam, S. The protein kinase complement of the human genome. Science 298, 1912–1934 (2002).
Nurse, P. Universal control mechanism regulating onset of M-phase. Nature 344, 503–508 (1990).
Pan, Z.Q., Amin, A. & Hurwitz, J. Characterization of the in vitro reconstituted cyclin A or B1-dependent cdk2 and cdc2 kinase activities. J. Biol. Chem. 268, 20443–20451 (1993).
Obenauer, J.C., Cantley, L.C. & Yaffe, M.B. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 31, 3635–3641 (2003).
Daile, P., Carnegie, P.R. & Young, J.D. Synthetic substrate for cyclic AMP-dependent protein kinase. Nature 257, 416–418 (1975).
Daile, P. & Carnegie, P.R. Peptides from myelin basic protein as substrates for adenosine 3′, 5′-cyclic monophosphate-dependent protein kinases. Biochem. Biophys. Res. Commun. 61, 852–858 (1974).
Kemp, B.E. & Pearson, R.B. Design and use of peptide substrates for protein kinases. Methods Enzymol. 200, 121–134 (1991).
Goldstein, D.M., Gray, N.S. & Zarrinkar, P.P. High-throughput kinase profiling as a platform for drug discovery. Nat. Rev. Drug Discov. 7, 391–397 (2008).
Schwartz, D. & Gygi, S.P. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 23, 1391–1398 (2005).
Manning, B.D. & Cantley, L.C. Hitting the target: emerging technologies in the search for kinase substrates. Sci. STKE 2002, PE49 (2002).
Parang, K., Kohn, J.A., Saldanha, S.A. & Cole, P.A. Development of photo-crosslinking reagents for protein kinase-substrate interactions. FEBS Lett. 520, 156–160 (2002).
Shen, K. & Cole, P.A. Conversion of a tyrosine kinase protein substrate to a high affinity ligand by ATP linkage. J. Am. Chem. Soc. 125, 16172–16173 (2003).
Linding, R. et al. Systematic discovery of in vivo phosphorylation networks. Cell 129, 1415–1426 (2007).
Johnson, S.A. & Hunter, T. Kinomics: methods for deciphering the kinome. Nat. Methods 2, 17–25 (2005).
Maly, D.J., Allen, J.A. & Shokat, K.M. A mechanism-based cross-linker for the identification of kinase-substrate pairs. J. Am. Chem. Soc. 126, 9160–9161 (2004).
Statsuk, A.V. et al. Tuning a three-component reaction for trapping kinase substrate complexes. J. Am. Chem. Soc. 130, 17568–17574 (2008).
Domon, B. & Aebersold, R. Mass spectrometry and protein analysis. Science 312, 212–217 (2006).
Andersen, J.S. et al. Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426, 570–574 (2003).
Andersen, J.S. et al. Nucleolar proteome dynamics. Nature 433, 77–83 (2005).
Foster, L.J. et al. A mammalian organelle map by protein correlation profiling. Cell 125, 187–199 (2006).
Rikova, K. et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131, 1190–1203 (2007).
Wilson-Grady, J.T., Villen, J. & Gygi, S.P. Phosphoproteome analysis of fission yeast. J. Proteome Res. 7, 1088–1097 (2008).
Zhai, B., Villen, J., Beausoleil, S.A., Mintseris, J. & Gygi, S.P. Phosphoproteome analysis of Drosophila melanogaster embryos. J. Proteome Res. 7, 1675–1682 (2008).
Dephoure, N. et al. A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. USA 105, 10762–10767 (2008).
Olsen, J.V. et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 (2006).
Elwood-Yen, K. et al. Inducible RNAi-Mediated Knock-down of PDK1 Fails to Prevent Tumor Formation in Multiple PTEN-Deficient Transgenic Mouse Models. American Association for Cancer Research Special Conference, Cambridge, MA, November 11–14, 2008.
Glass, D.B., Cheng, H.C., Mende-Mueller, L., Reed, J. & Walsh, D.A. Primary structural determinants essential for potent inhibition of cAMP-dependent protein kinase by inhibitory peptides corresponding to the active portion of the heat-stable inhibitor protein. J. Biol. Chem. 264, 8802–8810 (1989).
Villen, J. & Gygi, S.P. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat. Protocols 3, 1630–1638 (2008).
Rappsilber, J., Mann, M. & Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protocols 2, 1896–1906 (2007).
Wessel, D. & Flugge, U.I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143 (1984).
Haas, W. et al. Optimization and use of peptide mass measurement accuracy in shotgun proteomics. Mol. Cell. Proteomics 5, 1326–1337 (2006).
Kersey, P.J. et al. The International Protein Index: an integrated database for proteomics experiments. Proteomics 4, 1985–1988 (2004).
Eng, J.K., McCormack, A.L. & Yates, J.R. III. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 (1994).
Elias, J.E. & Gygi, S.P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 (2007).
Acknowledgements
This work was supported in part by grants from the National Institutes of Health (NIH; HG3456 and GM67945) and an industry-sponsored research project to S.P.G. from Merck and ThermoFisher. K.K. was supported by Daiichi Sankyo Co., Ltd. Y.Y. was partly supported by J. Blenis through grants from the NIH (GM51405). J.V. was supported by a grant from the Spanish Ministry of Education and Science. We thank C. Zhou, S. Elledge and N. Dephoure for help with cell cycle sample preparation, J. Elias and D. Kolippakkam for bioinformatics support, M. Rodriguez-Falcon for establishing Stagetip-IMAC protocol, and S. Sando and T. Robbins for peptide purification. We appreciate the advice and encouragement of many members of the Gygi lab and Mayumi Kubota.
Author information
Authors and Affiliations
Contributions
K.K. and R.A. are co-first authors. K.K., R.A., Y.Y., R.C.K., J.V. and S.P.G. participated in the planning, data generation and data interpretation. J.N.A., M.K., H.K., K.N., S.K., C.P. and R.C.H. prepared cell lysates using Akt and PDK1 inhibitors. A.S.F. and C.-L.W. prepared human renal carcinoma samples. J.R. synthesized all peptides. K.K. and S.P.G. wrote the manuscript and all authors edited it.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figs. 1–21, Supplementary Table 1, and Supplementary Notes (PDF 2047 kb)
Rights and permissions
About this article
Cite this article
Kubota, K., Anjum, R., Yu, Y. et al. Sensitive multiplexed analysis of kinase activities and activity-based kinase identification. Nat Biotechnol 27, 933–940 (2009). https://doi.org/10.1038/nbt.1566
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.1566
This article is cited by
-
Multiplex enzyme activity imaging by MALDI-IMS of substrate library conversions
Scientific Reports (2020)
-
Kinase inhibition profiles as a tool to identify kinases for specific phosphorylation sites
Nature Communications (2020)
-
Large-scale Discovery of Substrates of the Human Kinome
Scientific Reports (2019)
-
An ultrasensitive fiveplex activity assay for cellular kinases
Scientific Reports (2019)
-
Mapping phospho-catalytic dependencies of therapy-resistant tumours reveals actionable vulnerabilities
Nature Cell Biology (2019)