Abstract
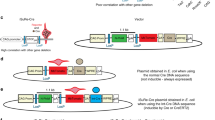
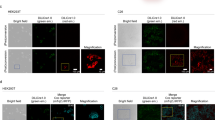
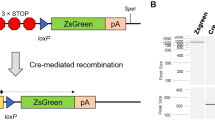
Functional redundancies, compensatory mechanisms, and lethal phenotypes often prevent the full analysis of gene functions through generation of germline null mutations in the mouse1. The use of site-specific recombinases, such as Cre, which catalyzes recombination between loxP sites2, has allowed the engineering of mice harboring targeted somatic mutations, which are both temporally controlled and cell-type restricted1,3. Many Cre-expressing mouse lines exist, but only a few transgenic lines are available that harbor a reporter gene whose expression is dependent on a Cre-mediated event3. Moreover, their use to monitor gene ablation at the level of individual cells is often limited, as in some tissues the reporter gene may be silenced1, be affected by position-effect variegation4, or reside in a chromatin configuration inaccessible for recombination5. Thus, one cannot validly extrapolate from the expression of a reporter transgene to an identical ablation pattern for the conditional allele of a given gene. By combining the ability of Cre recombinase to invert or excise a DNA fragment, depending on the orientation of the flanking loxP sites6, and the availability of both wild-type (WT) and mutant loxP sites7, we designed a Cre-dependent genetic switch (FLEx switch) through which the expression of a given gene is turned off, while the expression of another one is concomitantly turned on. We demonstrate the efficiency and reliability of this switch to readily detect, in the mouse, at the single cell level, Cre-mediated gene ablation. We discuss how this strategy can be used to generate genetic modifications in a conditional manner.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Metzger, D. & Chambon, P. Site- and time-specific gene targeting in the mouse. Methods 24, 71–80 (2001).
Sauer, B. & Henderson, N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc. Natl. Acad. Sci. USA 85, 5166–5170 (1988).
Nagy, A. Cre recombinase: the universal reagent for genome tailoring. Genesis 26, 99–109 (2000).
Montoliu, L., Chavez, S. & Vidal, M. Variegation associated with lacZ in transgenic animals: a warning note. Transgenic Res. 9, 237–239 (2000).
Vooijs, M., Jonkers, J. & Berns, A. A highly efficient ligand-regulated Cre recombinase mouse line shows that LoxP recombination is position dependent. EMBO Rep. 2, 292–297 (2001).
Abremski, K., Hoess, R. & Sternberg, N. Studies on the properties of P1 site-specific recombination: evidence for topologically unlinked products following recombination. Cell 32, 1301–1311 (1983).
Siegel, R.W., Jain, R. & Bradbury, A. Using an in vivo phagemid system to identify non-compatible loxP sequences. FEBS Lett. 499, 147–153 (2001).
Hoess, R.H., Wierzbicki, A. & Abremski, K. Formation of small circular DNA molecules via an in vitro site-specific recombination system. Gene 40, 325–329 (1985).
Ringrose, L. et al. Comparative kinetic analysis of FLP and cre recombinases: mathematical models for DNA binding and recombination. J. Mol. Biol. 284, 363–384 (1998).
Chapellier, B. et al. A conditional floxed (loxP-flanked) allele for the retinoic acid receptor gamma (RARγ) gene. Genesis 32, 95–98 (2002).
Lohnes, D. et al. Function of retinoic acid receptor gamma in the mouse. Cell 73, 643–658 (1993).
O' Gorman, S., Fox, D.T. & Wahl, G.M. Recombinase mediated gene activation and site-specific integration in mammalian cells. Science 251, 1351–1355 (1991).
Bonnerot, C., Rocancourt, D., Briand, P., Grimber, G. & Nicolas, J.F. A β-galactosidase hybrid protein targeted to nuclei as a marker for developmental studies. Proc. Natl. Acad. Sci. USA 84, 6795–6799 (1987).
Li, M. et al. RXR-α ablation in skin keratinocytes results in alopecia and epidermal alterations. Development 128, 675–688 (2000).
Lee, G. & Saito, I. Role of nucleotide sequences of loxP spacer region in Cre-mediated recombination. Gene 216, 55–65 (1998).
Kolb, A.F. Selection-marker-free modification of the murine β-casein gene using a lox2272 [correction of lox2722] site. Anal. Biochem. 290, 260–271 (2001).
Aranda, M. et al. Altered directionality in the Cre-LoxP site-specific recombination pathway. J. Mol. Biol. 311, 453–459 (2001).
Theis, M. et al. Endothelium-specific replacement of the connexin 43 coding region by a lacZ reporter gene. Genesis 29, 1–13 (2001).
Feng, Y.Q. et al. Site-specific chromosomal integration in mammalian cells: highly efficient Cre recombinase-mediated cassette exchange. J. Mol. Biol. 292, 779–785 (1999).
Buchholz, F., Angrand, P.O. & Stewart, F. A simple assay to determine the functionality of Cre or FLP recombination targets in genomic manipulation constructs. Nucleic Acids Res. 24, 3118–3119 (1996).
Acknowledgements
We thank B. Féret, G. Kimmich, and E. Blondelle for technical assistance, J.M. Garnier, A. Dierich, and M. Mark for their active contribution to this work, and G. Richards for critical reading of the manuscript. We also thank M. Li for K14-Cre mice and J.F. Nicolas for the lacZ gene. This work was supported by funds from the Centre National de la Recherche Scientifique (CNRS), the Institut National de la Santé et de la Recherche Médicale (INSERM), the Université Louis Pasteur (ULP), the Hôpital Universitaire de Strasbourg, the Collège de France, and the Association pour la Recherche sur le Cancer (ARC). F.S. was supported by a Marie Curie Fellowship from the European Community and C.C. by the Institut National de la Recherche Agronomique (INRA).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Schnütgen, F., Doerflinger, N., Calléja, C. et al. A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. Nat Biotechnol 21, 562–565 (2003). https://doi.org/10.1038/nbt811
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt811
This article is cited by
-
The structure of the NuA4–Tip60 complex reveals the mechanism and importance of long-range chromatin modification
Nature Structural & Molecular Biology (2023)
-
A blocking monoclonal antibody reveals dimerization of intracellular domains of ALK2 associated with genetic disorders
Nature Communications (2023)
-
Advanced neurobiological tools to interrogate metabolism
Nature Reviews Endocrinology (2023)
-
TCF7L2 acts as a molecular switch in midbrain to control mammal vocalization through its DNA binding domain but not transcription activation domain
Molecular Psychiatry (2023)
-
A new transgene mouse model using an extravesicular EGFP tag enables affinity isolation of cell-specific extracellular vesicles
Scientific Reports (2022)