Abstract
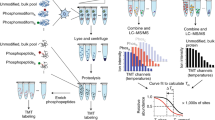
Protein phosphorylation is a dominant mechanism of information transfer in cells, and a major goal of current proteomic efforts is to generate a system-level map describing all the sites of protein phosphorylation. Recent efforts have focused on developing technologies for enriching and quantifying phosphopeptides. Identification of the sites of phosphorylation typically relies on tandem mass spectrometry to sequence individual peptides. Here we describe an approach for phosphopeptide mapping that makes it possible to interrogate a protein sequence directly with a protease that recognizes sites of phosphorylation. The key to this approach is the selective chemical transformation of phosphoserine and phosphothreonine residues into lysine analogs (aminoethylcysteine and β-methylaminoethylcysteine, respectively). Aminoethylcysteine-modified peptides are then cleaved with a lysine-specific protease to map sites of phosphorylation. A blocking step enables single-site cleavage, and adaptation of this reaction to the solid phase facilitates phosphopeptide enrichment and modification in one step.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Venter, J.C. et al. The sequence of the human genome. Science 291, 1304–1351 (2001).
Manning, G., Whyte, D.B., Martinez, R., Hunter, T. & Sudarsanam, S. The protein kinase complement of the human genome. Science 298, 1912–1934 (2002).
Shah, K. & Shokat, K.M. A chemical genetic screen for direct v-Src substrates reveals ordered assembly of a retrograde signaling pathway. Chem. Biol. 9, 35–47 (2002).
Bishop, A.C. et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407, 395–401 (2000).
McLachlin, D.T. & Chait, B.T. Analysis of phosphorylated proteins and peptides by mass spectrometry. Curr. Opin. Chem. Biol. 5, 591–602 (2001).
Mann, M. et al. Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol. 20, 261–268 (2002).
Zhou, H., Watts, J.D. & Aebersold, R. A systematic approach to the analysis of protein phosphorylation. Nat. Biotechnol. 19, 375–378 (2001).
Oda, Y., Nagasu, T. & Chait, B.T. Enrichment analysis of phosphorylated proteins as a tool for probing the phosphoproteome. Nat. Biotechnol. 19, 379–382 (2001).
Steen, H. & Mann, M. A new derivatization strategy for the analysis of phosphopeptides by precursor ion scanning in positive ion mode. J. Am. Soc. Mass Spectrom. 13, 996–1003 (2002).
Ficarro, S.B. et al. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 20, 301–305 (2002).
Meyer, H.E., Hoffmann-Posorske, E., Korte, H. & Heilmeyer, L.M. Jr. Sequence analysis of phosphoserine-containing peptides. Modification for picomolar sensitivity. FEBS Lett. 204, 61–66 (1986).
Simpson, D.L., Hranisavljevic, J. & Davidson, E.A. Elimination and sulfite addition as a means of localization and identification of substituted seryl and threonyl residues in proteins and proteoglycans. Biochemistry 11, 1849–1856 (1972).
Byford, M.F. Rapid and selective modification of phosphoserine residues catalysed by Ba2+ ions for their detection during peptide microsequencing. Biochem. J. 280 (Pt 1), 261–265 (1991).
Adamczyk, M., Gebler, J.C. & Wu, J. Selective analysis of phosphopeptides within a protein mixture by chemical modification, reversible biotinylation and mass spectrometry. Rapid Commun. Mass Spectrom. 15, 1481–1488 (2001).
Goshe, M.B. et al. Phosphoprotein isotope-coded affinity tag approach for isolating and quantitating phosphopeptides in proteome-wide analyses. Anal. Chem. 73, 2578–2586 (2001).
Jaffe, H., Veeranna & Pant, H.C. Characterization of serine and threonine phosphorylation sites in beta-elimination/ethanethiol addition-modified proteins by electrospray tandem mass spectrometry and database searching. Biochemistry 37, 16211–16224 (1998).
Annan, R.S., Huddleston, M.J., Verma, R., Deshaies, R.J. & Carr, S.A. A multidimensional electrospray MS-based approach to phosphopeptide mapping. Anal. Chem. 73, 393–404 (2001).
Janek, K., Wenschuh, H., Bienert, M. & Krause, E. Phosphopeptide analysis by positive and negative ion matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. 15, 1593–1599 (2001).
Limas, C.J. & Limas, C. Involvement of microtubules in the isoproterenol-induced 'down'-regulation of myocardial beta-adrenergic receptors. Biochim. Biophys. Acta 735, 181–184 (1983).
Haga, K., Ogawa, H., Haga, T. & Murofushi, H. GTP-binding-protein-coupled receptor kinase 2 (GRK2) binds and phosphorylates tubulin. Eur. J. Biochem. 255, 363–368 (1998).
Pitcher, J.A. et al. The G protein-coupled receptor kinase 2 is a microtubule-associated protein kinase that phosphorylates tubulin. J. Biol. Chem. 273, 12316–12324 (1998).
Carman, C.V., Som, T., Kim, C.M. & Benovic, J.L. Binding and phosphorylation of tubulin by G protein-coupled receptor kinases. J. Biol. Chem. 273, 20308–20316 (1998).
Banerjee, A. Coordination of posttranslational modifications of bovine brain alpha-tubulin. Polyglycylation of delta2 tubulin. J. Biol. Chem. 277, 46140–46144 (2002).
Alexander, J.E. et al. Characterization of posttranslational modifications in neuron-specific class III beta-tubulin by mass spectrometry. Proc. Natl. Acad. Sci. USA 88, 4685–4689 (1991).
Yoshida, N., Haga, K. & Haga, T. Identification of sites of phosphorylation by G-protein-coupled receptor kinase 2 in beta-tubulin. Eur. J. Biochem. 270, 1154–1163 (2003).
Pitcher, J.A. et al. Feedback inhibition of G protein-coupled receptor kinase 2 (GRK2) activity by extracellular signal-regulated kinases. J. Biol. Chem. 274, 34531–34534 (1999).
Beardsley, R.L., Karty, J.A. & Reilly, J.P. Enhancing the intensities of lysine-terminated tryptic peptide ions in matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. 14, 2147–2153 (2000).
Beardsley, R.L. & Reilly, J.P. Optimization of guanidination procedures for MALDI mass mapping. Anal. Chem. 74, 1884–1890 (2002).
Brancia, F.L., Oliver, S.G. & Gaskell, S.J. Improved matrix-assisted laser desorption/ionization mass spectrometric analysis of tryptic hydrolysates of proteins following guanidination of lysine-containing peptides. Rapid Commun. Mass Spectrom. 14, 2070–2073 (2000).
Cupo, P., El-Deiry, W., Whitney, P.L. & Awad, W.M. Jr. Stabilization of proteins by guanidination. J. Biol. Chem. 255, 10828–10833 (1980).
Kimmel, J.R. Guanidination of Proteins. Methods Enzymol. 11, 584–589 (1967).
Mega, T., Nakamura, N. & Ikenaka, T. Modifications of substituted seryl and threonyl residues in phosphopeptides and a polysialoglycoprotein by beta-elimination and nucleophile additions. J. Biochem. (Tokyo) 107, 68–72 (1990).
Wells, L.V. et al. Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Molecular and Cellular Proteomics 1, 791–804 (2002).
Greis, K.D. et al. Selective detection and site-analysis of O-GlcNAc-modified glycopeptides by beta-elimination and tandem electrospray mass spectrometry. Anal. Biochem. 234, 38–49 (1996).
Meldal, M. & Breddam, K. Anthranilamide and nitrotyrosine as a donor-acceptor pair in internally quenched fluorescent substrates for endopeptidases: multicolumn peptide synthesis of enzyme substrates for subtilisin Carlsberg and pepsin. Anal. Biochem. 195, 141–147 (1991).
Bonetto, V., Bergman, A.C., Jornvall, H. & Sillard, R. C-terminal sequence analysis of peptides and proteins using carboxypeptidases and mass spectrometry after derivatization of Lys and Cys residues. Anal. Chem. 69, 1315–1319 (1997).
Kim, C.M., Dion, S.B., Onorato, J.J. & Benovic, J.L. Expression and characterization of two beta-adrenergic receptor kinase isoforms using the baculovirus expression system. Receptor 3, 39–55 (1993).
Dorff, P.a.H. & Hauske, J.R. A Solid Phase CBZ Chloride Equivalent - A New Matrix Specific Linker. Tetrahedron Lett. 36, 1589–1592 (1995).
Acknowledgements
We thank Matt Simon and Dustin Maly for helpful advice. Z.A.K. is a Howard Hughes Medical Institute Predoctoral Fellow. K.M.S. acknowledges support from the National Institutes of Health (EB001987).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Knight, Z., Schilling, B., Row, R. et al. Phosphospecific proteolysis for mapping sites of protein phosphorylation. Nat Biotechnol 21, 1047–1054 (2003). https://doi.org/10.1038/nbt863
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt863
This article is cited by
-
Single-molecule mechanical fingerprinting with DNA nanoswitch calipers
Nature Nanotechnology (2021)
-
Current Trends in the Analysis of Post-translational Modifications
Chromatographia (2020)
-
Unblocking the Sink: Improved CID-Based Analysis of Phosphorylated Peptides by Enzymatic Removal of the BasicC-Terminal Residue
Journal of the American Society for Mass Spectrometry (2014)
-
Improving the Selectivity of the Phosphoric Acid β-Elimination on a Biotinylated Phosphopeptide
Journal of the American Society for Mass Spectrometry (2012)