Abstract
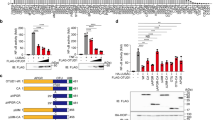
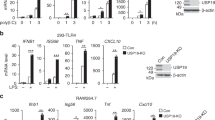
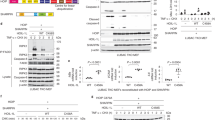
Nuclear factor-κB (NF-κB) is a key transcription factor in inflammatory, anti-apoptotic and immune processes. The ubiquitin pathway is crucial in regulating the NF-κB pathway. We have found that the LUBAC ligase complex, composed of the two RING finger proteins HOIL-1L and HOIP, conjugates a head-to-tail-linked linear polyubiquitin chain to substrates. Here, we demonstrate that LUBAC activates the canonical NF-κB pathway by binding to NEMO (NF-κB essential modulator, also called IKKγ) and conjugates linear polyubiquitin chains onto specific Lys residues in the CC2–LZ domain of NEMO in a Ubc13-independent manner. Moreover, in HOIL-1 knockout mice and cells derived from these mice, NF-κB signalling induced by pro-inflammatory cytokines such as TNF-α and IL-1β was suppressed, resulting in enhanced TNF-α–induced apoptosis in hepatocytes of HOIL-1 knockout mice. These results indicate that LUBAC is involved in the physiological regulation of the canonical NF-κB activation pathway through linear polyubiquitylation of NEMO.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Change history
13 January 2009
In the version of this article initially published online figure panels 3a, 3e and 7e were not clearly defined. Also (NEMOCC2~LZ) was corrected to (NEMO∆CC2~LZ). These errors have been corrected for the print, HTML and PDF versions of the article.
References
Hershko, A. & Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 (1998).
Kornitzer, D. & Ciechanover, A. Modes of regulation of ubiquitin-mediated protein degradation. J. Cell Physiol. 182, 1–11 (2000).
Hicke, L. & Dunn, R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 19, 141–172 (2003).
Chau, V. et al. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243, 1576–1583 (1989).
Deng, L. et al. Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103, 351–361 (2000).
Hofmann, R. M. & Pickart, C. M. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96, 645–653 (1999).
Kirisako, T. et al. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 25, 4877–4887 (2006).
Nakamura, M., Tokunaga, F., Sakata, S. & Iwai, K. Mutual regulation of conventional protein kinase C and a ubiquitin ligase complex. Biochem. Biophys. Res. Commun. 351, 340–347 (2006).
Hayden, M. S. & Ghosh, S. Signaling to NF-κB. Genes Dev. 18, 2195–2224 (2004).
Karin, M. & Greten, F. R. NF-κB: linking inflammation and immunity to cancer development and progression. Nature Rev. Immunol. 5, 749–759 (2005).
Chen, Z. J. Ubiquitin signalling in the NF-κB pathway. Nature Cell Biol. 7, 758–765 (2005).
Adhikari, A., Xu, M. & Chen, Z. J. Ubiquitin-mediated activation of TAK1 and IKK. Oncogene 26, 3214–3226 (2007).
Chen, Z. J., Parent, L. & Maniatis, T. Site-specific phosphorylation of IκBα by a novel ubiquitination-dependent protein kinase activity. Cell 84, 853–862 (1996).
Yamamoto, M. et al. Key function for the Ubc13 E2 ubiquitin-conjugating enzyme in immune receptor signaling. Nature Immunol. 7, 962–970 (2006).
Cong, Y. S., Yao, Y. L., Yang, W. M., Kuzhandaivelu, N. & Seto, E. The hepatitis B virus X-associated protein, XAP3, is a protein kinase C-binding protein. J. Biol. Chem. 272, 16482–16489 (1997).
Tokunaga, C. et al. Molecular cloning and characterization of a novel protein kinase C-interacting protein with structural motifs related to RBCC family proteins. Biochem. Biophys. Res. Commun. 244, 353–359 (1998).
Ea, C. K., Deng, L., Xia, Z. P., Pineda, G. & Chen, Z. J. Activation of IKK by TNFα requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol. Cell 22, 245–257 (2006).
Saito, N. et al. Two carboxyl-terminal activation regions of Epstein-Barr virus latent membrane protein 1 activate NF-κB through distinct signaling pathways in fibroblast cell lines. J. Biol. Chem. 278, 46565–46575 (2003).
Bubici, C., Papa, S., Pham, C. G., Zazzeroni, F. & Franzoso, G. NF-κB and JNK: an intricate affair. Cell Cycle 3, 1524–1529 (2004).
Papa, S. et al. The NF-κB-mediated control of the JNK cascade in the antagonism of programmed cell death in health and disease. Cell Death Differ. 13, 712–729 (2006).
Johnson, E. S., Ma, P. C., Ota, I. M. & Varshavsky, A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 270, 17442–17456 (1995).
Chen, F., Bhatia, D., Chang, Q. & Castranova, V. Finding NEMO by K63-linked polyubiquitin chain. Cell Death Differ. 13, 1835–1838 (2006).
Yamamoto, M. et al. Cutting Edge: Pivotal function of Ubc13 in thymocyte TCR signaling. J. Immunol. 177, 7520–7524 (2006).
Abbott, D. W., Wilkins, A., Asara, J. M. & Cantley, L. C. The Crohn's disease protein, NOD2, requires RIP2 in order to induce ubiquitinylation of a novel site on NEMO. Curr. Biol. 14, 2217–2227 (2004).
Sebban-Benin, H. et al. Identification of TRAF6-dependent NEMO polyubiquitination sites through analysis of a new NEMO mutation causing incontinentia pigmenti. Hum. Mol. Genet. 16, 2805–2815 (2007).
Huang, T. T., Wuerzberger-Davis, S. M., Wu, Z. H. & Miyamoto, S. Sequential modification of NEMO/IKKγ by SUMO-1 and ubiquitin mediates NF-κB activation by genotoxic stress. Cell 115, 565–576 (2003).
Newton, K. et al. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell 134, 668–678 (2008).
Brummelkamp, T. R., Nijman, S. M., Dirac, A. M. & Bernards, R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-κB. Nature 424, 797–801 (2003).
Evans, P. C. et al. Zinc-finger protein A20, a regulator of inflammation and cell survival, has de-ubiquitinating activity. Biochem. J. 378, 727–734 (2004).
Dworkin-Rastl, E., Shrutkowski, A. & Dworkin, M. B. Multiple ubiquitin mRNAs during Xenopus laevis development contain tandem repeats of the 76 amino acid coding sequence. Cell 39, 321–325 (1984).
Einspanier, R., Sharma, H. S. & Scheit, K. H. Cloning and sequence analysis of a cDNA encoding poly-ubiquitin in human ovarian granulosa cells. Biochem. Biophys. Res. Commun. 147, 581–587 (1987).
Kim, J. H., Park, K. C., Chung, S. S., Bang, O. & Chung, C. H. Deubiquitinating enzymes as cellular regulators. J. Biochem. 134, 9–18 (2003).
Ventii, K. H. et al. Protein partners of deubiquitinating enzymes. Biochem. J. 414, 161–175 (2008).
Karin, M. Nuclear factor-κB in cancer development and progression. Nature 441, 431–436 (2006).
Braun, T. et al. Targeting NF-κB in hematologic malignancies. Cell Death Differ. 13, 748–758 (2006).
Janssens, S. & Tschopp, J. Signals from within: the DNA-damage-induced NF-κB response. Cell Death Differ. 13, 773–784 (2006).
Thompson, H. G., Harris, J. W., Lin, L. & Brody, J. P. Identification of the protein Zibra, its genomic organization, regulation, and expression in breast cancer cells. Exp. Cell Res. 295, 448–459 (2004).
Swerdlow, P. S., Finley, D. & Varshavsky, A. Enhancement of immunoblot sensitivity by heating of hydrated filters. Anal. Biochem. 156, 147–153 (1986).
Iwai, K. et al. Identification of the von Hippel-lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc. Natl Acad. Sci. USA 96, 12436–12441 (1999).
Fernandez-de-Cossio, J. et al. Automated interpretation of low-energy collision-induced dissociation spectra by SeqMS, a software aid for de novo sequencing by tandem mass spectrometry. Electrophoresis 21, 1694–1699 (2000).
Acknowledgements
We would like to thank Nobuhito Goda and Natsuko Fujiki for instruction in mouse primary hepatocyte preparation; Ayumi Taya for supporting the LC and MS analyses; and Koichi Nakajima for reagents. This work was partly supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (F.T., T.T. and K.I.), the Japan Health Sciences Foundation (F.T.) and the Naito Foundation (F.T.).
Author information
Authors and Affiliations
Contributions
F.T., S.-I.S, K.K., T.N. and M.K. performed experiments; Y.S., Y.S. and T.T. conducted mass spectrometric analyses. S.-I.S. and S.M. generated the HOIL-1 KO mice; T.K., S.Y., M.Y., S.A. and K.T. provided reagents and suggestions; F.T. and K.I. coordinated the study and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1915 kb)
Rights and permissions
About this article
Cite this article
Tokunaga, F., Sakata, Si., Saeki, Y. et al. Involvement of linear polyubiquitylation of NEMO in NF-κB activation. Nat Cell Biol 11, 123–132 (2009). https://doi.org/10.1038/ncb1821
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb1821
This article is cited by
-
LUBAC-mediated M1 Ub regulates necroptosis by segregating the cellular distribution of active MLKL
Cell Death & Disease (2024)
-
LUBAC promotes angiogenesis and lung tumorigenesis by ubiquitinating and antagonizing autophagic degradation of HIF1α
Oncogenesis (2024)
-
LUBAC is required for RIG-I sensing of RNA viruses
Cell Death & Differentiation (2024)
-
RNF31 promotes proliferation and invasion of hepatocellular carcinoma via nuclear factor kappaB activation
Scientific Reports (2024)
-
Mechanisms underlying linear ubiquitination and implications in tumorigenesis and drug discovery
Cell Communication and Signaling (2023)