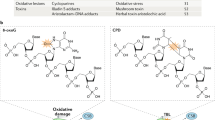
Abstract
Most mammalian cells in nature are quiescent but actively transcribing mRNA for normal physiological processes; thus, it is important to investigate how endogenous and exogenous DNA damage compromises transcription in cells. Here we describe a new competitive transcription and adduct bypass (CTAB) assay to determine the effects of DNA lesions on the fidelity and efficiency of transcription. Using this strategy, we demonstrate that the oxidatively induced lesions 8,5′-cyclo-2′-deoxyadenosine (cdA) and 8,5′-cyclo-2′-deoxyguanosine (cdG) and the methylglyoxal-induced lesion N2-(1-carboxyethyl)-2′-deoxyguanosine (N2-CEdG) strongly inhibited transcription in vitro and in mammalian cells. In addition, cdA and cdG, but not N2-CEdG, induced transcriptional mutagenesis in vitro and in vivo. Furthermore, when located on the template DNA strand, all examined lesions were primarily repaired by transcription-coupled nucleotide excision repair in mammalian cells. This newly developed CTAB assay should be generally applicable for quantitatively assessing how other DNA lesions affect DNA transcription in vitro and in cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Brégeon, D. & Doetsch, P.W. Transcriptional mutagenesis: causes and involvement in tumour development. Nat. Rev. Cancer 11, 218–227 (2011).
Lindahl, T. Instability and decay of the primary structure of DNA. Nature 362, 709–715 (1993).
Saxowsky, T.T. & Doetsch, P.W. RNA polymerase encounters with DNA damage: transcription-coupled repair or transcriptional mutagenesis? Chem. Rev. 106, 474–488 (2006).
Hanawalt, P.C. & Spivak, G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 9, 958–970 (2008).
Kuraoka, I. & Tanaka, K. Assays for transcription elongation by RNA polymerase II using oligo(dC)-tailed template with single DNA damage. Methods Enzymol. 408, 214–223 (2006).
Saxowsky, T.T., Meadows, K.L., Klungland, A. & Doetsch, P.W. 8-Oxoguanine-mediated transcriptional mutagenesis causes Ras activation in mammalian cells. Proc. Natl. Acad. Sci. USA 105, 18877–18882 (2008).
Brégeon, D., Peignon, P.A. & Sarasin, A. Transcriptional mutagenesis induced by 8-oxoguanine in mammalian cells. PLoS Genet. 5, e1000577 (2009).
Burns, J.A., Dreij, K., Cartularo, L. & Scicchitano, D.A. O6-methylguanine induces altered proteins at the level of transcription in human cells. Nucleic Acids Res. 38, 8178–8187 (2010).
Brégeon, D., Doddridge, Z.A., You, H.J., Weiss, B. & Doetsch, P.W. Transcriptional mutagenesis induced by uracil and 8-oxoguanine in Escherichia coli. Mol. Cell 12, 959–970 (2003).
Viswanathan, A., You, H.J. & Doetsch, P.W. Phenotypic change caused by transcriptional bypass of uracil in nondividing cells. Science 284, 159–162 (1999).
Brooks, P.J. et al. The oxidative DNA lesion 8,5′-(S)-cyclo-2′-deoxyadenosine is repaired by the nucleotide excision repair pathway and blocks gene expression in mammalian cells. J. Biol. Chem. 275, 22355–22362 (2000).
Marietta, C. & Brooks, P.J. Transcriptional bypass of bulky DNA lesions causes new mutant RNA transcripts in human cells. EMBO Rep. 8, 388–393 (2007).
Kuraoka, I. et al. Removal of oxygen free-radical-induced 5′,8-purine cyclodeoxynucleosides from DNA by the nucleotide excision-repair pathway in human cells. Proc. Natl. Acad. Sci. USA 97, 3832–3837 (2000).
Jaruga, P. & Dizdaroglu, M. 8,5′-Cyclopurine-2′-deoxynucleosides in DNA: mechanisms of formation, measurement, repair and biological effects. DNA Repair (Amst.) 7, 1413–1425 (2008).
Belmadoui, N. et al. Radiation-induced formation of purine 5′,8-cyclonucleosides in isolated and cellular DNA: high stereospecificity and modulating effect of oxygen. Org. Biomol. Chem. 8, 3211–3219 (2010).
Chatgilialoglu, C., Ferreri, C. & Terzidis, M.A. Purine 5′,8-cyclonucleoside lesions: chemistry and biology. Chem. Soc. Rev. 40, 1368–1382 (2011).
Wang, J. et al. Quantification of oxidative DNA lesions in tissues of Long-Evans Cinnamon rats by capillary high-performance liquid chromatography-tandem mass spectrometry coupled with stable isotope-dilution method. Anal. Chem. 83, 2201–2209 (2011).
D'Errico, M. et al. New functions of XPC in the protection of human skin cells from oxidative damage. EMBO J. 25, 4305–4315 (2006).
Rodriguez, H. et al. Lymphoblasts of women with BRCA1 mutations are deficient in cellular repair of 8,5′-cyclopurine-2′-deoxynucleosides and 8-hydroxy-2′-deoxyguanosine. Biochemistry 46, 2488–2496 (2007).
Thornalley, P.J. Pharmacology of methylglyoxal: formation, modification of proteins and nucleic acids, and enzymatic detoxification—a role in pathogenesis and antiproliferative chemotherapy. Gen. Pharmacol. 27, 565–573 (1996).
Frischmann, M., Bidmon, C., Angerer, J. & Pischetsrieder, M. Identification of DNA adducts of methylglyoxal. Chem. Res. Toxicol. 18, 1586–1592 (2005).
Schneider, M. et al. Determination of glycated nucleobases in human urine by a new monoclonal antibody specific for N2-carboxyethyl-2′-deoxyguanosine. Chem. Res. Toxicol. 17, 1385–1390 (2004).
Li, H. et al. N2-carboxyethyl-2′-deoxyguanosine, a DNA glycation marker, in kidneys and aortas of diabetic and uremic patients. Kidney Int. 69, 388–392 (2006).
Synold, T. et al. Advanced glycation end products of DNA: quantification of N2-(1-carboxyethyl)-2′-deoxyguanosine in biological samples by liquid chromatography electrospray ionization tandem mass spectrometry. Chem. Res. Toxicol. 21, 2148–2155 (2008).
Yuan, B., Cao, H., Jiang, Y., Hong, H. & Wang, Y. Efficient and accurate bypass of N2-(1-carboxyethyl)-2′-deoxyguanosine by DinB DNA polymerase in vitro and in vivo. Proc. Natl. Acad. Sci. USA 105, 8679–8684 (2008).
Murata-Kamiya, N., Kaji, H. & Kasai, H. Deficient nucleotide excision repair increases base-pair substitutions but decreases TGGC frameshifts induced by methylglyoxal in Escherichia coli. Mutat. Res. 442, 19–28 (1999).
Tamae, D., Lim, P., Wuenschell, G.E. & Termini, J. Mutagenesis and repair induced by the DNA advanced glycation end product N2-1-(carboxyethyl)-2′-deoxyguanosine in human cells. Biochemistry 50, 2321–2329 (2011).
Delaney, J.C. & Essigmann, J.M. Mutagenesis, genotoxicity, and repair of 1-methyladenine, 3-alkylcytosines, 1-methylguanine, and 3-methylthymine in alkB Escherichia coli. Proc. Natl. Acad. Sci. USA 101, 14051–14056 (2004).
Delaney, J.C. & Essigmann, J.M. Assays for determining lesion bypass efficiency and mutagenicity of site-specific DNA lesions in vivo. Methods Enzymol. 408, 1–15 (2006).
Masters, B.S., Stohl, L.L. & Clayton, D.A. Yeast mitochondrial RNA polymerase is homologous to those encoded by bacteriophages T3 and T7. Cell 51, 89–99 (1987).
Yuan, B. et al. The roles of DNA polymerases κ and ι in the error-free bypass of N2-carboxyalkyl-2′-deoxyguanosine lesions in mammalian cells. J. Biol. Chem. 286, 17503–17511 (2011).
Hong, H., Cao, H. & Wang, Y. Formation and genotoxicity of a guanine-cytosine intrastrand cross-link lesion in vivo. Nucleic Acids Res. 35, 7118–7127 (2007).
Rolig, R.L. et al. Survival, mutagenesis, and host cell reactivation in a Chinese hamster ovary cell ERCC1 knock-out mutant. Mutagenesis 12, 277–283 (1997).
Yuan, B., Wang, J., Cao, H., Sun, R. & Wang, Y. High-throughput analysis of the mutagenic and cytotoxic properties of DNA lesions by next-generation sequencing. Nucleic Acids Res. 39, 5945–5954 (2011).
Clauson, C.L., Oestreich, K.J., Austin, J.W. & Doetsch, P.W. Abasic sites and strand breaks in DNA cause transcriptional mutagenesis in Escherichia coli. Proc. Natl. Acad. Sci. USA 107, 3657–3662 (2010).
Clauson, C.L., Saxowsky, T.T. & Doetsch, P.W. Dynamic flexibility of DNA repair pathways in growth arrested Escherichia coli. DNA Repair (Amst.) 9, 842–847 (2010).
Jasti, V.P. et al. (5′S)-8,5′-Cyclo-2′-deoxyguanosine is a strong block to replication, a potent pol V-dependent mutagenic lesion, and is inefficiently repaired in Escherichia coli. Biochemistry 50, 3862–3865 (2011).
Kuraoka, I. et al. Oxygen free radical damage to DNA. Translesion synthesis by human DNA polymerase η and resistance to exonuclease action at cyclopurine deoxynucleoside residues. J. Biol. Chem. 276, 49283–49288 (2001).
Tornaletti, S. Transcription arrest at DNA damage sites. Mutat. Res. 577, 131–145 (2005).
Steitz, T.A. The structural changes of T7 RNA polymerase from transcription initiation to elongation. Curr. Opin. Struct. Biol. 19, 683–690 (2009).
Kornberg, R. The molecular basis of eukaryotic transcription (Nobel Lecture). Angew. Chem. Int. Edn Engl. 46, 6956–6965 (2007).
Brooks, P.J. The 8,5′-cyclopurine-2′-deoxynucleosides: candidate neurodegenerative DNA lesions in xeroderma pigmentosum, and unique probes of transcription and nucleotide excision repair. DNA Repair (Amst.) 7, 1168–1179 (2008).
Choi, J.Y. & Guengerich, F.P. Kinetic evidence for inefficient and error-prone bypass across bulky N2-guanine DNA adducts by human DNA polymerase ι. J. Biol. Chem. 281, 12315–12324 (2006).
Choi, J.Y., Angel, K.C. & Guengerich, F.P. Translesion synthesis across bulky N2-alkyl guanine DNA adducts by human DNA polymerase κ. J. Biol. Chem. 281, 21062–21072 (2006).
Jarosz, D.F., Godoy, V.G., Delaney, J.C., Essigmann, J.M. & Walker, G.C. A single amino acid governs enhanced activity of DinB DNA polymerases on damaged templates. Nature 439, 225–228 (2006).
Cohen, S.E. et al. Roles for the transcription elongation factor NusA in both DNA repair and damage tolerance pathways in Escherichia coli. Proc. Natl. Acad. Sci. USA 107, 15517–15522 (2010).
Cheng, T.F., Hu, X., Gnatt, A. & Brooks, P.J. Differential blocking effects of the acetaldehyde-derived DNA lesion N2-ethyl-2′-deoxyguanosine on transcription by multisubunit and single subunit RNA polymerases. J. Biol. Chem. 283, 27820–27828 (2008).
Acknowledgements
We thank T.R. O'Connor (City of Hope), G.P. Pfeifer (City of Hope) and M. Seidman (National Institute of Aging) for providing cell lines and plasmid. This work was supported by the US National Institutes of Health (R01 DK082779, R01 ES019873 and R01 CA101864 to Y.W. and R01 ES016114 to L.J.N.).
Author information
Authors and Affiliations
Contributions
C.Y., X.D., B.Y. and Y.W. designed research; C.Y., X.D., B.Y. and Jianshuang Wang performed research; C.Y., X.D. and Y.W. analyzed data; C.Y., Jin Wang, P.J.B., L.J.N. and Y.W. wrote the paper; Y.W. conceived and supervised the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Methods and Supplementary Results (PDF 1646 kb)
Rights and permissions
About this article
Cite this article
You, C., Dai, X., Yuan, B. et al. A quantitative assay for assessing the effects of DNA lesions on transcription. Nat Chem Biol 8, 817–822 (2012). https://doi.org/10.1038/nchembio.1046
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1046
This article is cited by
-
Quantitative measurement of transcriptional inhibition and mutagenesis induced by site-specifically incorporated DNA lesions in vitro and in vivo
Nature Protocols (2015)
-
Identification of DNA lesions using a third base pair for amplification and nanopore sequencing
Nature Communications (2015)
-
Effects of Tet-mediated Oxidation Products of 5-Methylcytosine on DNA Transcription in vitro and in Mammalian Cells
Scientific Reports (2014)
-
A selective USP1–UAF1 inhibitor links deubiquitination to DNA damage responses
Nature Chemical Biology (2014)