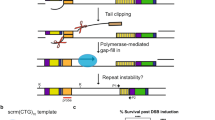
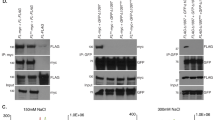
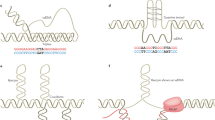
Abstract
Instability of (CTG)•(CAG) microsatellite trinucleotide repeat (TNR) sequences is responsible for more than a dozen neurological or neuromuscular diseases. TNR instability during DNA synthesis is thought to involve slipped-strand or hairpin structures in template or nascent DNA strands, although direct evidence for hairpin formation in human cells is lacking. We have used targeted recombination to create a series of isogenic HeLa cell lines in which (CTG)•(CAG) repeats are replicated from an ectopic copy of the Myc (also known as c-myc) replication origin. In this system, the tendency of chromosomal (CTG)•(CAG) tracts to expand or contract was affected by origin location and the leading or lagging strand replication orientation of the repeats, and instability was enhanced by prolonged cell culture, increased TNR length and replication inhibition. Hairpin cleavage by synthetic zinc finger nucleases in these cells has provided the first direct evidence for the formation of hairpin structures during replication in vivo.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Pearson, C.E., Edamura, K.N. & Cleary, J.D. Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 6, 729–742 (2005).
Lia, A.S. et al. Somatic instability of the CTG repeat in mice transgenic for the myotonic dystrophy region is age dependent but not correlated to the relative intertissue transcription levels and proliferative capacities. Hum. Mol. Genet. 7, 1285–1291 (1998).
Fortune, M.T., Vassilopoulos, C., Coolbaugh, M.I., Siciliano, M.J. & Monckton, D.G. Dramatic, expansion-biased, age-dependent, tissue-specific somatic mosaicism in a transgenic mouse model of triplet repeat instability. Hum. Mol. Genet. 9, 439–445 (2000).
Miret, J.J., Pessoa-Brandao, L. & Lahue, R.S. Orientation-dependent and sequence-specific expansions of CTG/CAG trinucleotide repeats in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 95, 12438–12443 (1998).
Petruska, J., Arnheim, N. & Goodman, M.F. Stability of intrastrand hairpin structures formed by the CAG/CTG class of DNA triplet repeats associated with neurological diseases. Nucleic Acids Res. 24, 1992–1998 (1996).
Pearson, C.E., Wang, Y.H., Griffith, J.D. & Sinden, R.R. Structural analysis of slipped-strand DNA (S-DNA) formed in (CTG)n(CAG)n repeats from the myotonic dystrophy locus. Nucleic Acids Res. 26, 816–823 (1998).
Pelletier, R., Krasilnikova, M.M., Samadashwily, G.M., Lahue, R. & Mirkin, S.M. Replication and expansion of trinucleotide repeats in yeast. Mol. Cell. Biol. 23, 1349–1357 (2003).
McMurray, C.T. DNA secondary structure: a common and causative factor for expansion in human disease. Proc. Natl. Acad. Sci. USA 96, 1823–1825 (1999).
Mirkin, S.M. Expandable DNA repeats and human disease. Nature 447, 932–940 (2007).
Sinden, R.R., Pytlos, M.J. & Potaman, V. Mechanisms of DNA repeat expansion. in Human Nucleotide Expansion Disorders (eds. Fry, M. & Usdin, K.) 3–53 (Springer, Berlin; New York, 2006).
Hashem, V.I. et al. Chemotherapeutic deletion of CTG repeats in lymphoblast cells from DM1 patients. Nucleic Acids Res. 32, 6334–6346 (2004).
Yang, Z., Lau, R., Marcadier, J.L., Chitayat, D. & Pearson, C.E. Replication inhibitors modulate instability of an expanded trinucleotide repeat at the myotonic dystrophy type 1 disease locus in human cells. Am. J. Hum. Genet. 73, 1092–1105 (2003).
Cleary, J.D. & Pearson, C.E. Replication fork dynamics and dynamic mutations: the fork-shift model of repeat instability. Trends Genet. 21, 272–280 (2005).
Trinh, T.Q. & Sinden, R.R. Preferential DNA secondary structure mutagenesis in the lagging strand of replication in E. coli. Nature 352, 544–547 (1991).
Yang, J. & Freudenreich, C.H. Haploinsufficiency of yeast FEN1 causes instability of expanded CAG/CTG tracts in a length-dependent manner. Gene 393, 110–115 (2007).
Spiro, C. & McMurray, C.T. Nuclease-deficient FEN-1 blocks Rad51/BRCA1-mediated repair and causes trinucleotide repeat instability. Mol. Cell. Biol. 23, 6063–6074 (2003).
van den Broek, W.J., Nelen, M.R., van der Heijden, G.W., Wansink, D.G. & Wieringa, B. Fen1 does not control somatic hypermutability of the (CTG)(n)•(CAG)(n) repeat in a knock-in mouse model for DM1. FEBS Lett. 580, 5208–5214 (2006).
Mirkin, E.V. & Mirkin, S.M. Replication fork stalling at natural impediments. Microbiol. Mol. Biol. Rev. 71, 13–35 (2007).
Delagoutte, E., Goellner, G.M., Guo, J., Baldacci, G. & McMurray, C.T. Single-stranded DNA-binding protein in vitro eliminates the orientation-dependent impediment to polymerase passage on CAG/CTG repeats. J. Biol. Chem. 283, 13341–13356 (2008).
Malott, M. & Leffak, M. Activity of the c-myc replicator at an ectopic chromosomal location. Mol. Cell. Biol. 19, 5685–5695 (1999).
Liu, G., Malott, M. & Leffak, M. Multiple functional elements comprise a mammalian chromosomal replicator. Mol. Cell. Biol. 23, 1832–1842 (2003).
Liu, G., Bissler, J.J., Sinden, R.R. & Leffak, M. Unstable spinocerebellar ataxia Type 10 (ATTCT)•(AGAAT) repeats are associated with aberrant replication at the ATX10 locus and replication origin-dependent expansion at an ectopic site in human cells. Mol. Cell. Biol. 27, 7828–7838 (2007).
Burhans, W.C. et al. Emetine allows identification of origins of mammalian DNA replication by imbalanced DNA synthesis, not through conservative nucleosome segregation. EMBO J. 10, 4351–4360 (1991).
Gacy, A.M. & McMurray, C.T. Influence of hairpins on template reannealing at trinucleotide repeat duplexes: a model for slipped DNA. Biochemistry 37, 9426–9434 (1998).
Bitinaite, J., Wah, D.A., Aggarwal, A.K. & Schildkraut, I. FokI dimerization is required for DNA cleavage. Proc. Natl. Acad. Sci. USA 95, 10570–10575 (1998).
Mani, M., Smith, J., Kandavelou, K., Berg, J.M. & Chandrasegaran, S. Binding of two zinc finger nuclease monomers to two specific sites is required for effective double-strand DNA cleavage. Biochem. Biophys. Res. Commun. 334, 1191–1197 (2005).
Segal, D.J., Dreier, B., Beerli, R.R. & Barbas, C.F. III. Toward controlling gene expression at will: selection and design of zinc finger domains recognizing each of the 5′-GNN-3′ DNA target sequences. Proc. Natl. Acad. Sci. USA 96, 2758–2763 (1999).
Mittelman, D. et al. Zinc-finger directed double-strand breaks within CAG repeat tracts promote repeat instability in human cells. Proc. Natl. Acad. Sci. USA 106, 9607–9612 (2009).
Carroll, D., Morton, J.J., Beumer, K.J. & Segal, D.J. Design, construction and in vitro testing of zinc finger nucleases. Nat. Protoc. 1, 1329–1341 (2006).
Usdin, K. & Woodford, K.J. CGG repeats associated with DNA instability and chromosome fragility form structures that block DNA synthesis in vitro. Nucleic Acids Res. 23, 4202–4209 (1995).
Freudenreich, C.H., Kantrow, S.M. & Zakian, V.A. Expansion and length-dependent fragility of CTG repeats in yeast. Science 279, 853–856 (1998).
Carbajal Rivera, M.L., Ordaz Tellez, M.G., Castaneda Sortibran, A. & Rodriguez-Arnaiz, R. Emetine and/or its metabolites are genotoxic in somatic cells of Drosophila melanogaster. J. Toxicol. Environ. Health A 70, 1713–1716 (2007).
Hashem, V.I., Rosche, W.A. & Sinden, R.R. Genetic recombination destabilizes (CTG)n.(CAG)n repeats in E. coli. Mutat. Res. 554, 95–109 (2004).
Mirkin, S.M. DNA structures, repeat expansions and human hereditary disorders. Curr. Opin. Struct. Biol. 16, 351–358 (2006).
Gordenin, D.A., Kunkel, T.A. & Resnick, M.A. Repeat expansion–all in a flap? Nat. Genet. 16, 116–118 (1997).
Lin, Y., Dion, V. & Wilson, J.H. Transcription promotes contraction of CAG repeat tracts in human cells. Nat. Struct. Mol. Biol. 13, 179–180 (2006).
Jackson, S.M. et al. A SCA7 CAG/CTG repeat expansion is stable in Drosophila melanogaster despite modulation of genomic context and gene dosage. Gene 347, 35–41 (2005).
Libby, R.T. et al. Genomic context drives SCA7 CAG repeat instability, while expressed SCA7 cDNAs are intergenerationally and somatically stable in transgenic mice. Hum. Mol. Genet. 12, 41–50 (2003).
Monckton, D.G., Wong, L.J., Ashizawa, T. & Caskey, C.T. Somatic mosaicism, germline expansions, germline reversions and intergenerational reductions in myotonic dystrophy males: small pool PCR analyses. Hum. Mol. Genet. 4, 1–8 (1995).
Seznec, H. et al. Transgenic mice carrying large human genomic sequences with expanded CTG repeat mimic closely the DM CTG repeat intergenerational and somatic instability. Hum. Mol. Genet. 9, 1185–1194 (2000).
Khajavi, M. et al. “Mitotic drive” of expanded CTG repeats in myotonic dystrophy type 1 (DM1). Hum. Mol. Genet. 10, 855–863 (2001).
Hashem, V.I., Rosche, W.A. & Sinden, R.R. Genetic assays for measuring rates of (CAG).(CTG) repeat instability in Escherichia coli. Mutat. Res. 502, 25–37 (2002).
Shishkin, A.A. et al. Large-scale expansions of Friedreich's ataxia GAA repeats in yeast. Mol. Cell 35, 82–92 (2009).
Bidichandani, S.I., Ashizawa, T. & Patel, P.I. The GAA triplet-repeat expansion in Friedreich ataxia interferes with transcription and may be associated with an unusual DNA structure. Am. J. Hum. Genet. 62, 111–121 (1998).
Potaman, V.N. et al. Unpaired Structures in SCA10 (ATTCT)(n)•(AGAAT)(n) Repeats. J. Mol. Biol. 326, 1095–1111 (2003).
Sinden, R.R., Pytlos-Sinden, M.J. & Potaman, V.N. Slipped strand DNA structures. Front. Biosci. 12, 4788–4799 (2007).
Voineagu, I., Surka, C.F., Shishkin, A.A., Krasilnikova, M.M. & Mirkin, S.M. Replisome stalling and stabilization at CGG repeats, which are responsible for chromosomal fragility. Nat. Struct. Mol. Biol. 16, 226–228 (2009).
Sander, J.D., Zaback, P., Joung, J.K., Voytas, D.F. & Dobbs, D. Zinc Finger Targeter (ZiFiT): an engineered zinc finger/target site design tool. Nucleic Acids Res. 35, W599–W605 (2007).
Acknowledgements
The authors thank C. Pearson and D.G. Monckton for their comments on this work. This work was supported by a grant from the Wright State University Boonshoft School of Medicine to G.L. and by grants from the US National Institutes of Health to M.L. (GM53819) and to J.J.B. (DK61458).
Author information
Authors and Affiliations
Contributions
Experiments were conceived and designed by G.L., M.L., J.J.B. and R.R.S. and performed by G.L., X.C. and M.L. The manuscript was drafted by G.L. and M.L. and revised by all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–11 (PDF 3709 kb)
Rights and permissions
About this article
Cite this article
Liu, G., Chen, X., Bissler, J. et al. Replication-dependent instability at (CTG)•(CAG) repeat hairpins in human cells. Nat Chem Biol 6, 652–659 (2010). https://doi.org/10.1038/nchembio.416
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.416
This article is cited by
-
Dynamic alternative DNA structures in biology and disease
Nature Reviews Genetics (2023)
-
Precise CAG repeat contraction in a Huntington’s Disease mouse model is enabled by gene editing with SpCas9-NG
Communications Biology (2021)
-
Huntington disease: new insights into molecular pathogenesis and therapeutic opportunities
Nature Reviews Neurology (2020)
-
Break-induced replication sparks CGG-repeat instability
Nature Structural & Molecular Biology (2018)
-
Mechanisms of genetic instability caused by (CGG)n repeats in an experimental mammalian system
Nature Structural & Molecular Biology (2018)